Gene
KWMTBOMO05257
Pre Gene Modal
BGIBMGA002417
Annotation
PREDICTED:_UPF0415_protein_C7orf25_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.809
Sequence
CDS
ATGTCGAAAATCAATCATGCTGAAGATTTAGTGGAAGTGGTTAAAGATAAAATCAAGTTTGGTGAAAATTTAATAATACAGCTACAACCAATTCAGCATGTTGAAGGTGTTATGAAATTGCAAAGGAAAATTAAGCAAGAAATAGAATTTTTGAAGAGGTTGGAAAAATCAAAAAACTTCAAGATTGAACAGTTGGCTTGTTCTAACCTGCGTCATCTTGGATCAATGGTGGAGTGCGCGCTTAGACCTTATGTGCTCGCTGTGTGTAAAACATTTCACATAGATAACTGTAATAAACTAATAATAGACATTGTTGGGGATCAAGGCAATACATGGACCAAAGTGATAGCAAGAAATCCAAAATCTTTATCTGCTTTAAGTGCAGGGAAATCTTCATATGGAGCAAGATCTATTTTAGATCAAGCTGATGACTACTTAAAGTGCTCTAGGCTATACCCATGCATGTACCAGCCACCAAAGGTTGTGTTTGAATTTATGAGTGGAATAGAAGAAAATTTAGCAAATAAATTAAGAGCAGTTGGTATTGTTGTGAAGGGTGAAATTTTACCAAATTCAATTGTTCAAGACAGTGGTGGTGAATCCTTTGATTCCCAAAGCAGTGATGATGAATCAAATTGCATTAATATGCAATCAAAGATACCTGAGCTTAATCATGGCATAGATCATTCTGAAATAAAAACTCTAAACTTGGATGTAACAACCATGATGGCATATGTGAGCAACATGACAAATGGCCATCACAATTTTATCTTTAAACAGGAAGTTTTGACACAGCAATGCTCTTGGGAAGCAGAGAGACCTGTAAAACCAATACTAAGAAAATTATTTGAAGGCAAGAAACTAGTCTGCTGTGAGACAGCATGGAATGACTTTGAAAAAATACTTGAAACATTAGGAGGACCAATGGAAAAGAAAAGAACTGTTCTTTTAAAAGAAATGATTACAGTTTATCCTGATGACTTTGGAGGTGAGGATGATTATCCGAGGCAAAATCTTAAAGTTCGAGGCCACGTTCGAGTAAGATCCAAGATTATATTCAATTTTGGTCATCGAATCAAAGCGCTTACAGTCTCGGCAAACGAAGGCTTCGTCAGGGCTGCTCTTCAACAGGGCGTAACATATGCAGCTTTTGTTCACGAGTCAAGAGCTCTCACTGAAGGGAAGGAAGACTCTGCAAGAAGGATAACTTTATGA
Protein
MSKINHAEDLVEVVKDKIKFGENLIIQLQPIQHVEGVMKLQRKIKQEIEFLKRLEKSKNFKIEQLACSNLRHLGSMVECALRPYVLAVCKTFHIDNCNKLIIDIVGDQGNTWTKVIARNPKSLSALSAGKSSYGARSILDQADDYLKCSRLYPCMYQPPKVVFEFMSGIEENLANKLRAVGIVVKGEILPNSIVQDSGGESFDSQSSDDESNCINMQSKIPELNHGIDHSEIKTLNLDVTTMMAYVSNMTNGHHNFIFKQEVLTQQCSWEAERPVKPILRKLFEGKKLVCCETAWNDFEKILETLGGPMEKKRTVLLKEMITVYPDDFGGEDDYPRQNLKVRGHVRVRSKIIFNFGHRIKALTVSANEGFVRAALQQGVTYAAFVHESRALTEGKEDSARRITL
Summary
Uniprot
H9IYT2
A0A194RNC5
A0A194QBU9
A0A2H1WJG2
A0A3S2P7W9
A0A212F1E5
+ More
A0A1W4WUU1 A0A0T6B4R1 A0A1B6C4V1 A0A2J7QUU5 A0A067QNK2 A0A1Y1NM92 A0A1W4X4C8 K7JUF5 U4UGT1 A0A139WM07 A0A182TDK0 A0A182VE45 A0A182XIF0 A0A182KZF7 Q7PHB7 W5JF31 A0A182IRY6 Q16V90 A0A182HYS8 A0A182F911 A0A182QE23 A0A182W3J7 A0A182RE06 A0A182PHX3 A0A182Y8P0 A0A2M4AVL8 A0A182N9W1 A0A182M5A5 A0A182T7E4 A0A195DN12 A0A182JR89 A0A151WZ76 A0A034WFB3 F4W4J0 A0A195EZJ8 A0A0K8TMW2 A0A087ZSD2 A0A195BIM9 A0A158P342 A0A0A1XFK5 A0A2A3EPG4 A0A0C9S0M2 E9IIL4 A0A182GW52 A0A195CCT8 E2C3J9 W8BLX3 A0A182GKB5 A0A0K8WK66 A0A1Q3F0P0 T1JKY6 A0A1I8PVU7 A0A154P9R4 A0A026W6T4 B0W0V8 A0A232F6S8 A0A0L7RCC1 A0A1I8NGG6 A0A1B6JV62 B4J2Q9 E0VSR9 E2A1D0 B4LGJ0 A0A0L0C0M4 B3M8E8 B4N4B5 B4L0C4 A0A0M5J827 A0A0P4VPM6 R4G4C4 B3NJC1 A0A1B0A5B3 A0A1A9XLY4 A0A1A9X5N5 A0A1B0FML4 A0A0V0G9C7 B4HVK1 B4QLF6 Q9W0M6 Q8IGH5 B4PCN5 A0A3L8E2J8 A0A023F7Y4 A0A3B0K451 A0A087T062 A0A224XS79 A0A146M6L3 A0A1W4W0L5 A0A0A9XSZ0 Q2LZ81 A0A1A9UT16 A0A293MUQ7
A0A1W4WUU1 A0A0T6B4R1 A0A1B6C4V1 A0A2J7QUU5 A0A067QNK2 A0A1Y1NM92 A0A1W4X4C8 K7JUF5 U4UGT1 A0A139WM07 A0A182TDK0 A0A182VE45 A0A182XIF0 A0A182KZF7 Q7PHB7 W5JF31 A0A182IRY6 Q16V90 A0A182HYS8 A0A182F911 A0A182QE23 A0A182W3J7 A0A182RE06 A0A182PHX3 A0A182Y8P0 A0A2M4AVL8 A0A182N9W1 A0A182M5A5 A0A182T7E4 A0A195DN12 A0A182JR89 A0A151WZ76 A0A034WFB3 F4W4J0 A0A195EZJ8 A0A0K8TMW2 A0A087ZSD2 A0A195BIM9 A0A158P342 A0A0A1XFK5 A0A2A3EPG4 A0A0C9S0M2 E9IIL4 A0A182GW52 A0A195CCT8 E2C3J9 W8BLX3 A0A182GKB5 A0A0K8WK66 A0A1Q3F0P0 T1JKY6 A0A1I8PVU7 A0A154P9R4 A0A026W6T4 B0W0V8 A0A232F6S8 A0A0L7RCC1 A0A1I8NGG6 A0A1B6JV62 B4J2Q9 E0VSR9 E2A1D0 B4LGJ0 A0A0L0C0M4 B3M8E8 B4N4B5 B4L0C4 A0A0M5J827 A0A0P4VPM6 R4G4C4 B3NJC1 A0A1B0A5B3 A0A1A9XLY4 A0A1A9X5N5 A0A1B0FML4 A0A0V0G9C7 B4HVK1 B4QLF6 Q9W0M6 Q8IGH5 B4PCN5 A0A3L8E2J8 A0A023F7Y4 A0A3B0K451 A0A087T062 A0A224XS79 A0A146M6L3 A0A1W4W0L5 A0A0A9XSZ0 Q2LZ81 A0A1A9UT16 A0A293MUQ7
Pubmed
19121390
26354079
22118469
24845553
28004739
20075255
+ More
23537049 18362917 19820115 20966253 12364791 14747013 17210077 20920257 23761445 17510324 25244985 25348373 21719571 26369729 21347285 25830018 21282665 26483478 20798317 24495485 24508170 28648823 25315136 17994087 20566863 26108605 27129103 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 25474469 26823975 25401762 15632085
23537049 18362917 19820115 20966253 12364791 14747013 17210077 20920257 23761445 17510324 25244985 25348373 21719571 26369729 21347285 25830018 21282665 26483478 20798317 24495485 24508170 28648823 25315136 17994087 20566863 26108605 27129103 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 25474469 26823975 25401762 15632085
EMBL
BABH01010861
BABH01010862
KQ459984
KPJ19012.1
KQ459220
KPJ02894.1
+ More
ODYU01009018 SOQ53106.1 RSAL01000222 RVE44015.1 AGBW02010898 OWR47565.1 LJIG01009900 KRT82155.1 GEDC01030266 GEDC01028884 JAS07032.1 JAS08414.1 NEVH01010480 PNF32358.1 KK853123 KDR11077.1 GEZM01002049 JAV97396.1 KB632314 ERL92237.1 KQ971317 KYB28972.1 AAAB01008898 EAA44604.4 ADMH02001667 ETN61495.1 CH477599 EAT38478.1 APCN01005404 AXCN02001202 GGFK01011499 MBW44820.1 AXCM01000198 KQ980713 KYN14295.1 KQ982649 KYQ53067.1 GAKP01005558 JAC53394.1 GL887532 EGI70885.1 KQ981905 KYN33551.1 GDAI01001919 JAI15684.1 KQ976467 KYM84145.1 ADTU01007725 GBXI01004103 JAD10189.1 KZ288204 PBC33152.1 GBYB01010084 GBYB01014126 JAG79851.1 JAG83893.1 GL763494 EFZ19562.1 JXUM01092549 JXUM01092550 JXUM01092551 JXUM01092552 JXUM01092553 KQ563994 KXJ72962.1 KQ977935 KYM98674.1 GL452320 EFN77537.1 GAMC01004466 GAMC01004465 JAC02091.1 JXUM01069668 JXUM01069669 KQ562563 KXJ75609.1 GDHF01026431 GDHF01001079 JAI25883.1 JAI51235.1 GFDL01013946 JAV21099.1 JH431430 KQ434853 KZC08635.1 KK107447 EZA50739.1 DS231819 EDS42775.1 NNAY01000829 OXU26292.1 KQ414615 KOC68642.1 GECU01024593 GECU01004616 JAS83113.1 JAT03091.1 CH916366 EDV96050.1 AAZO01005175 DS235756 EEB16425.1 GL435760 EFN72756.1 CH940647 EDW69428.1 JRES01001065 KNC25863.1 CH902618 EDV38883.1 CH964095 EDW78989.1 CH933809 EDW18070.2 CP012525 ALC43536.1 GDKW01001646 JAI54949.1 ACPB03008663 GAHY01001490 JAA76020.1 CH954178 EDV50083.1 CCAG010017555 GECL01001619 JAP04505.1 CH480817 EDW49966.1 CM000363 CM002912 EDX08729.1 KMY96723.1 AE014296 BT128838 AAF47417.2 AEM92669.1 BT001782 AAN71537.1 CM000159 EDW92758.1 QOIP01000001 RLU26960.1 GBBI01001383 JAC17329.1 OUUW01000012 SPP87482.1 KK112767 KFM58501.1 GFTR01005575 JAW10851.1 GDHC01003957 GDHC01002601 JAQ14672.1 JAQ16028.1 GBHO01019762 GBHO01019761 GBRD01014683 JAG23842.1 JAG23843.1 JAG51143.1 CH379069 EAL29627.3 GFWV01019794 MAA44522.1
ODYU01009018 SOQ53106.1 RSAL01000222 RVE44015.1 AGBW02010898 OWR47565.1 LJIG01009900 KRT82155.1 GEDC01030266 GEDC01028884 JAS07032.1 JAS08414.1 NEVH01010480 PNF32358.1 KK853123 KDR11077.1 GEZM01002049 JAV97396.1 KB632314 ERL92237.1 KQ971317 KYB28972.1 AAAB01008898 EAA44604.4 ADMH02001667 ETN61495.1 CH477599 EAT38478.1 APCN01005404 AXCN02001202 GGFK01011499 MBW44820.1 AXCM01000198 KQ980713 KYN14295.1 KQ982649 KYQ53067.1 GAKP01005558 JAC53394.1 GL887532 EGI70885.1 KQ981905 KYN33551.1 GDAI01001919 JAI15684.1 KQ976467 KYM84145.1 ADTU01007725 GBXI01004103 JAD10189.1 KZ288204 PBC33152.1 GBYB01010084 GBYB01014126 JAG79851.1 JAG83893.1 GL763494 EFZ19562.1 JXUM01092549 JXUM01092550 JXUM01092551 JXUM01092552 JXUM01092553 KQ563994 KXJ72962.1 KQ977935 KYM98674.1 GL452320 EFN77537.1 GAMC01004466 GAMC01004465 JAC02091.1 JXUM01069668 JXUM01069669 KQ562563 KXJ75609.1 GDHF01026431 GDHF01001079 JAI25883.1 JAI51235.1 GFDL01013946 JAV21099.1 JH431430 KQ434853 KZC08635.1 KK107447 EZA50739.1 DS231819 EDS42775.1 NNAY01000829 OXU26292.1 KQ414615 KOC68642.1 GECU01024593 GECU01004616 JAS83113.1 JAT03091.1 CH916366 EDV96050.1 AAZO01005175 DS235756 EEB16425.1 GL435760 EFN72756.1 CH940647 EDW69428.1 JRES01001065 KNC25863.1 CH902618 EDV38883.1 CH964095 EDW78989.1 CH933809 EDW18070.2 CP012525 ALC43536.1 GDKW01001646 JAI54949.1 ACPB03008663 GAHY01001490 JAA76020.1 CH954178 EDV50083.1 CCAG010017555 GECL01001619 JAP04505.1 CH480817 EDW49966.1 CM000363 CM002912 EDX08729.1 KMY96723.1 AE014296 BT128838 AAF47417.2 AEM92669.1 BT001782 AAN71537.1 CM000159 EDW92758.1 QOIP01000001 RLU26960.1 GBBI01001383 JAC17329.1 OUUW01000012 SPP87482.1 KK112767 KFM58501.1 GFTR01005575 JAW10851.1 GDHC01003957 GDHC01002601 JAQ14672.1 JAQ16028.1 GBHO01019762 GBHO01019761 GBRD01014683 JAG23842.1 JAG23843.1 JAG51143.1 CH379069 EAL29627.3 GFWV01019794 MAA44522.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000192223
+ More
UP000235965 UP000027135 UP000002358 UP000030742 UP000007266 UP000075902 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075880 UP000008820 UP000075840 UP000069272 UP000075886 UP000075920 UP000075900 UP000075885 UP000076408 UP000075884 UP000075883 UP000075901 UP000078492 UP000075881 UP000075809 UP000007755 UP000078541 UP000005203 UP000078540 UP000005205 UP000242457 UP000069940 UP000249989 UP000078542 UP000008237 UP000095300 UP000076502 UP000053097 UP000002320 UP000215335 UP000053825 UP000095301 UP000001070 UP000009046 UP000000311 UP000008792 UP000037069 UP000007801 UP000007798 UP000009192 UP000092553 UP000015103 UP000008711 UP000092445 UP000092443 UP000091820 UP000092444 UP000001292 UP000000304 UP000000803 UP000002282 UP000279307 UP000268350 UP000054359 UP000192221 UP000001819 UP000078200
UP000235965 UP000027135 UP000002358 UP000030742 UP000007266 UP000075902 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075880 UP000008820 UP000075840 UP000069272 UP000075886 UP000075920 UP000075900 UP000075885 UP000076408 UP000075884 UP000075883 UP000075901 UP000078492 UP000075881 UP000075809 UP000007755 UP000078541 UP000005203 UP000078540 UP000005205 UP000242457 UP000069940 UP000249989 UP000078542 UP000008237 UP000095300 UP000076502 UP000053097 UP000002320 UP000215335 UP000053825 UP000095301 UP000001070 UP000009046 UP000000311 UP000008792 UP000037069 UP000007801 UP000007798 UP000009192 UP000092553 UP000015103 UP000008711 UP000092445 UP000092443 UP000091820 UP000092444 UP000001292 UP000000304 UP000000803 UP000002282 UP000279307 UP000268350 UP000054359 UP000192221 UP000001819 UP000078200
Pfam
Interpro
IPR041076
DUF5614
+ More
IPR010733 DUF1308
IPR001650 Helicase_C
IPR001098 DNA-dir_DNA_pol_A_palm_dom
IPR036390 WH_DNA-bd_sf
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR002298 DNA_polymerase_A
IPR019760 DNA-dir_DNA_pol_A_CS
IPR027417 P-loop_NTPase
IPR011017 TRASH_dom
IPR000988 Ribosomal_L24e-rel
IPR038630 L24e/L24_sf
IPR039540 UBL3-like_ubiquitin_dom
IPR029071 Ubiquitin-like_domsf
IPR010733 DUF1308
IPR001650 Helicase_C
IPR001098 DNA-dir_DNA_pol_A_palm_dom
IPR036390 WH_DNA-bd_sf
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR002298 DNA_polymerase_A
IPR019760 DNA-dir_DNA_pol_A_CS
IPR027417 P-loop_NTPase
IPR011017 TRASH_dom
IPR000988 Ribosomal_L24e-rel
IPR038630 L24e/L24_sf
IPR039540 UBL3-like_ubiquitin_dom
IPR029071 Ubiquitin-like_domsf
Gene 3D
ProteinModelPortal
H9IYT2
A0A194RNC5
A0A194QBU9
A0A2H1WJG2
A0A3S2P7W9
A0A212F1E5
+ More
A0A1W4WUU1 A0A0T6B4R1 A0A1B6C4V1 A0A2J7QUU5 A0A067QNK2 A0A1Y1NM92 A0A1W4X4C8 K7JUF5 U4UGT1 A0A139WM07 A0A182TDK0 A0A182VE45 A0A182XIF0 A0A182KZF7 Q7PHB7 W5JF31 A0A182IRY6 Q16V90 A0A182HYS8 A0A182F911 A0A182QE23 A0A182W3J7 A0A182RE06 A0A182PHX3 A0A182Y8P0 A0A2M4AVL8 A0A182N9W1 A0A182M5A5 A0A182T7E4 A0A195DN12 A0A182JR89 A0A151WZ76 A0A034WFB3 F4W4J0 A0A195EZJ8 A0A0K8TMW2 A0A087ZSD2 A0A195BIM9 A0A158P342 A0A0A1XFK5 A0A2A3EPG4 A0A0C9S0M2 E9IIL4 A0A182GW52 A0A195CCT8 E2C3J9 W8BLX3 A0A182GKB5 A0A0K8WK66 A0A1Q3F0P0 T1JKY6 A0A1I8PVU7 A0A154P9R4 A0A026W6T4 B0W0V8 A0A232F6S8 A0A0L7RCC1 A0A1I8NGG6 A0A1B6JV62 B4J2Q9 E0VSR9 E2A1D0 B4LGJ0 A0A0L0C0M4 B3M8E8 B4N4B5 B4L0C4 A0A0M5J827 A0A0P4VPM6 R4G4C4 B3NJC1 A0A1B0A5B3 A0A1A9XLY4 A0A1A9X5N5 A0A1B0FML4 A0A0V0G9C7 B4HVK1 B4QLF6 Q9W0M6 Q8IGH5 B4PCN5 A0A3L8E2J8 A0A023F7Y4 A0A3B0K451 A0A087T062 A0A224XS79 A0A146M6L3 A0A1W4W0L5 A0A0A9XSZ0 Q2LZ81 A0A1A9UT16 A0A293MUQ7
A0A1W4WUU1 A0A0T6B4R1 A0A1B6C4V1 A0A2J7QUU5 A0A067QNK2 A0A1Y1NM92 A0A1W4X4C8 K7JUF5 U4UGT1 A0A139WM07 A0A182TDK0 A0A182VE45 A0A182XIF0 A0A182KZF7 Q7PHB7 W5JF31 A0A182IRY6 Q16V90 A0A182HYS8 A0A182F911 A0A182QE23 A0A182W3J7 A0A182RE06 A0A182PHX3 A0A182Y8P0 A0A2M4AVL8 A0A182N9W1 A0A182M5A5 A0A182T7E4 A0A195DN12 A0A182JR89 A0A151WZ76 A0A034WFB3 F4W4J0 A0A195EZJ8 A0A0K8TMW2 A0A087ZSD2 A0A195BIM9 A0A158P342 A0A0A1XFK5 A0A2A3EPG4 A0A0C9S0M2 E9IIL4 A0A182GW52 A0A195CCT8 E2C3J9 W8BLX3 A0A182GKB5 A0A0K8WK66 A0A1Q3F0P0 T1JKY6 A0A1I8PVU7 A0A154P9R4 A0A026W6T4 B0W0V8 A0A232F6S8 A0A0L7RCC1 A0A1I8NGG6 A0A1B6JV62 B4J2Q9 E0VSR9 E2A1D0 B4LGJ0 A0A0L0C0M4 B3M8E8 B4N4B5 B4L0C4 A0A0M5J827 A0A0P4VPM6 R4G4C4 B3NJC1 A0A1B0A5B3 A0A1A9XLY4 A0A1A9X5N5 A0A1B0FML4 A0A0V0G9C7 B4HVK1 B4QLF6 Q9W0M6 Q8IGH5 B4PCN5 A0A3L8E2J8 A0A023F7Y4 A0A3B0K451 A0A087T062 A0A224XS79 A0A146M6L3 A0A1W4W0L5 A0A0A9XSZ0 Q2LZ81 A0A1A9UT16 A0A293MUQ7
Ontologies
PANTHER
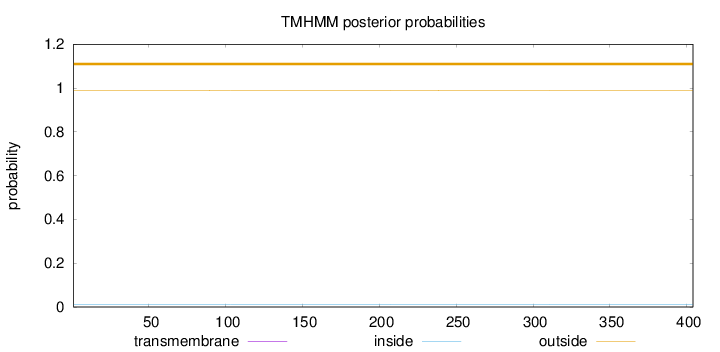
Topology
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00256
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.01044
outside
1 - 404
Population Genetic Test Statistics
Pi
261.279962
Theta
189.893672
Tajima's D
1.402625
CLR
0
CSRT
0.75721213939303
Interpretation
Uncertain
