Pre Gene Modal
BGIBMGA002492
Annotation
PREDICTED:_RWD_domain-containing_protein_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.683 ER Reliability : 1.663 Nuclear Reliability : 1.257
Sequence
CDS
ATGGATTACAATTACGAACAAACTAGTGAAGTGGAAGCATTAGATTCTATTTATTTCGGCGATATGACAGTGATAGAAACAAAACCATTTCACAAATTTAGTATACCTATAAAGTCAGAGGGGTTTGATGATGGAGAGGGGTTGGCCTGCCATCTAGTATTTACTTACACATCAAAGTATCCTGATGAATTACCCATTATTGAAATTGACAATGAGGAGAATTTCGACAATATTGTTGATAAAGAGGAACTTCTCACACATCTTGCTGAACAGGGTAAAGAAAACTTGGGAATGGTCATGGTATTCACATTAGTGTCAGCTGGACAAGAGTGGCTTAATGAGACTTGGGATAAAGTTAAGAAAGAGCGTGAAGAAAGGATACTAGCCAAACAAAAGGCAGATGAAGAGGCAGAACTGAAAAGGTTTGAAGGCACTAGAGTCACAGTAGAGTCATTTTTGGCATGGAGGAAACAGTTTGAGATTGACATGGGAATCCCAGCTAAGAGGGAACGAGAAGCAAAAGATAAGAACAAACTGACTGGAAGAGAACTATTCCTTCAGGATACAACACTCAATGAATCGGATCTCAAATTCTTGGATGATGGAGATGCAGTAAAAGTGGATGAGTCTTTATTCCAGAACCTGGATGATTTGGAACTCACCGATGAGGATGATGGTGACTATGAACCTTCACAAAGTGGTAGTGATAGTGATTAG
Protein
MDYNYEQTSEVEALDSIYFGDMTVIETKPFHKFSIPIKSEGFDDGEGLACHLVFTYTSKYPDELPIIEIDNEENFDNIVDKEELLTHLAEQGKENLGMVMVFTLVSAGQEWLNETWDKVKKEREERILAKQKADEEAELKRFEGTRVTVESFLAWRKQFEIDMGIPAKREREAKDKNKLTGRELFLQDTTLNESDLKFLDDGDAVKVDESLFQNLDDLELTDEDDGDYEPSQSGSDSD
Summary
Uniprot
H9IZ07
A0A3S2LCY7
A0A1E1W2J7
A0A194QD36
A0A2H1WP44
A0A194RNM0
+ More
A0A212EN86 S4PA54 A0A1Y1MMN2 K7J2C8 A0A232FBB7 A0A0C9QKD3 J3JYQ6 A0A0T6BFK8 D6X4V8 A0A0K8TQ58 E2AB33 E9IHP2 A0A195D463 A0A151WEY8 E2BB57 A0A067QVT5 A0A026WTQ7 A0A158NW01 A0A0L0BN32 A0A2J7QK69 A0A195BN19 A0A195FYC5 A0A1I8NTN3 A0A336MZ64 T1PM15 A0A151J3N9 B4NAG7 A0A1A9WEE5 A0A1B6L7S6 A0A2S2QWP0 A0A1B6II37 B4JH42 A0A1A9XL80 A0A1B0B8E0 A0A182K0K9 A0A182N6K3 A0A154PKV0 A0A088AFK5 A0A1L8DCU4 A0A182GU83 A0A182QLP4 A0A1L8DCE8 A0A1B0GAF7 A0A1A9ZUP8 A0A1A9VGG4 A0A1B6G4M2 A0A023EKW0 A0A182TYR1 A0A182LC08 Q7QBP1 A0A182HIW6 A0A182HD58 A0A182USQ4 A0A182X8B1 A0A2J7QK63 A0A182FPK0 A0A2M4BYE7 W5JA47 A0A2M4BY75 A0A2M3Z8I9 J9JMP3 A0A182XVR5 C4WTU9 A0A182RXZ8 B4MBX2 A0A2M4AHU3 A0A2S2NM15 A0A034WBQ2 A0A1W4WE68 A0A182SQ10 T1PFA3 A0A0M9AAE2 A0A182P2Y6 A0A182MU80 A0A0M3QXF4 A0A0L7R4A4 A0A182W0B3 A0A182IVR8 B4K7F4 Q171H7 F4WN22 B3M0K5 A0A1B0GN99 A0A1B6DR73 W8BNT0 B4G3Z9 Q298I8 B4PL00 B3P776 Q9VCB9 B4QSI3 B4HGJ9 A0A1B0CKR5 A0A084WS75
A0A212EN86 S4PA54 A0A1Y1MMN2 K7J2C8 A0A232FBB7 A0A0C9QKD3 J3JYQ6 A0A0T6BFK8 D6X4V8 A0A0K8TQ58 E2AB33 E9IHP2 A0A195D463 A0A151WEY8 E2BB57 A0A067QVT5 A0A026WTQ7 A0A158NW01 A0A0L0BN32 A0A2J7QK69 A0A195BN19 A0A195FYC5 A0A1I8NTN3 A0A336MZ64 T1PM15 A0A151J3N9 B4NAG7 A0A1A9WEE5 A0A1B6L7S6 A0A2S2QWP0 A0A1B6II37 B4JH42 A0A1A9XL80 A0A1B0B8E0 A0A182K0K9 A0A182N6K3 A0A154PKV0 A0A088AFK5 A0A1L8DCU4 A0A182GU83 A0A182QLP4 A0A1L8DCE8 A0A1B0GAF7 A0A1A9ZUP8 A0A1A9VGG4 A0A1B6G4M2 A0A023EKW0 A0A182TYR1 A0A182LC08 Q7QBP1 A0A182HIW6 A0A182HD58 A0A182USQ4 A0A182X8B1 A0A2J7QK63 A0A182FPK0 A0A2M4BYE7 W5JA47 A0A2M4BY75 A0A2M3Z8I9 J9JMP3 A0A182XVR5 C4WTU9 A0A182RXZ8 B4MBX2 A0A2M4AHU3 A0A2S2NM15 A0A034WBQ2 A0A1W4WE68 A0A182SQ10 T1PFA3 A0A0M9AAE2 A0A182P2Y6 A0A182MU80 A0A0M3QXF4 A0A0L7R4A4 A0A182W0B3 A0A182IVR8 B4K7F4 Q171H7 F4WN22 B3M0K5 A0A1B0GN99 A0A1B6DR73 W8BNT0 B4G3Z9 Q298I8 B4PL00 B3P776 Q9VCB9 B4QSI3 B4HGJ9 A0A1B0CKR5 A0A084WS75
Pubmed
19121390
26354079
22118469
23622113
28004739
20075255
+ More
28648823 22516182 23537049 18362917 19820115 26369729 20798317 21282665 24845553 24508170 30249741 21347285 26108605 25315136 17994087 26483478 24945155 20966253 12364791 14747013 17210077 20920257 23761445 25244985 25348373 17510324 21719571 24495485 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588
28648823 22516182 23537049 18362917 19820115 26369729 20798317 21282665 24845553 24508170 30249741 21347285 26108605 25315136 17994087 26483478 24945155 20966253 12364791 14747013 17210077 20920257 23761445 25244985 25348373 17510324 21719571 24495485 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588
EMBL
BABH01010853
RSAL01000222
RVE44011.1
GDQN01011506
GDQN01009854
GDQN01004827
+ More
GDQN01000197 JAT79548.1 JAT81200.1 JAT86227.1 JAT90857.1 KQ459220 KPJ02890.1 ODYU01010028 SOQ54843.1 KQ459984 KPJ19009.1 AGBW02013684 OWR42965.1 GAIX01003399 JAA89161.1 GEZM01026840 GEZM01026839 JAV86932.1 AAZX01001570 NNAY01000566 OXU27627.1 GBYB01004024 JAG73791.1 APGK01055167 BT128386 KB741261 KB632344 AEE63344.1 ENN71583.1 ERL93040.1 LJIG01000891 KRT86049.1 KQ971381 EEZ97231.1 GDAI01001081 JAI16522.1 GL438234 EFN69309.1 GL763289 EFZ19905.1 KQ976870 KYN07695.1 KQ983238 KYQ46377.1 GL446916 EFN87078.1 KK852898 KDR14162.1 KK107109 QOIP01000008 EZA59343.1 RLU19162.1 ADTU01027600 JRES01001706 KNC20689.1 NEVH01013277 PNF28977.1 KQ976433 KYM87383.1 KQ981169 KYN45307.1 UFQT01002505 UFQT01003386 SSX33592.1 SSX34951.1 KA649647 KA649769 AFP64398.1 KQ980265 KYN17059.1 CH964232 EDW80781.1 GEBQ01020198 JAT19779.1 GGMS01012727 MBY81930.1 GECU01021112 GECU01004071 JAS86594.1 JAT03636.1 CH916369 EDV92733.1 JXJN01009913 KQ434948 KZC12486.1 GFDF01009944 JAV04140.1 JXUM01018606 KQ560517 KXJ82024.1 AXCN02000287 GFDF01009946 JAV04138.1 CCAG010015110 GECZ01012379 JAS57390.1 GAPW01003765 JAC09833.1 AAAB01008879 EAA08430.4 APCN01002095 JXUM01034045 KQ561010 KXJ80047.1 PNF28975.1 GGFJ01008893 MBW58034.1 ADMH02001712 ETN61332.1 GGFJ01008894 MBW58035.1 GGFM01004070 MBW24821.1 ABLF02036049 AK340777 BAH71319.1 CH940656 EDW58593.1 GGFK01007038 MBW40359.1 GGMR01005612 MBY18231.1 GAKP01007235 JAC51717.1 KA646845 AFP61474.1 KQ435710 KOX79756.1 AXCM01000475 CP012526 ALC45833.1 KQ414657 KOC65715.1 CH933806 EDW14278.1 CH477455 EAT40639.1 GL888233 EGI64390.1 CH902617 EDV44252.1 AJVK01028559 GEDC01009110 JAS28188.1 GAMC01006078 JAC00478.1 CH479179 EDW24410.1 CM000070 EAL27967.1 CM000160 EDW98985.1 CH954182 EDV53896.1 AE014297 BT099549 AAF56253.1 ACU32633.1 CM000364 EDX14182.1 CH480815 EDW43452.1 AJWK01016549 AJWK01016550 ATLV01026348 KE525410 KFB53069.1
GDQN01000197 JAT79548.1 JAT81200.1 JAT86227.1 JAT90857.1 KQ459220 KPJ02890.1 ODYU01010028 SOQ54843.1 KQ459984 KPJ19009.1 AGBW02013684 OWR42965.1 GAIX01003399 JAA89161.1 GEZM01026840 GEZM01026839 JAV86932.1 AAZX01001570 NNAY01000566 OXU27627.1 GBYB01004024 JAG73791.1 APGK01055167 BT128386 KB741261 KB632344 AEE63344.1 ENN71583.1 ERL93040.1 LJIG01000891 KRT86049.1 KQ971381 EEZ97231.1 GDAI01001081 JAI16522.1 GL438234 EFN69309.1 GL763289 EFZ19905.1 KQ976870 KYN07695.1 KQ983238 KYQ46377.1 GL446916 EFN87078.1 KK852898 KDR14162.1 KK107109 QOIP01000008 EZA59343.1 RLU19162.1 ADTU01027600 JRES01001706 KNC20689.1 NEVH01013277 PNF28977.1 KQ976433 KYM87383.1 KQ981169 KYN45307.1 UFQT01002505 UFQT01003386 SSX33592.1 SSX34951.1 KA649647 KA649769 AFP64398.1 KQ980265 KYN17059.1 CH964232 EDW80781.1 GEBQ01020198 JAT19779.1 GGMS01012727 MBY81930.1 GECU01021112 GECU01004071 JAS86594.1 JAT03636.1 CH916369 EDV92733.1 JXJN01009913 KQ434948 KZC12486.1 GFDF01009944 JAV04140.1 JXUM01018606 KQ560517 KXJ82024.1 AXCN02000287 GFDF01009946 JAV04138.1 CCAG010015110 GECZ01012379 JAS57390.1 GAPW01003765 JAC09833.1 AAAB01008879 EAA08430.4 APCN01002095 JXUM01034045 KQ561010 KXJ80047.1 PNF28975.1 GGFJ01008893 MBW58034.1 ADMH02001712 ETN61332.1 GGFJ01008894 MBW58035.1 GGFM01004070 MBW24821.1 ABLF02036049 AK340777 BAH71319.1 CH940656 EDW58593.1 GGFK01007038 MBW40359.1 GGMR01005612 MBY18231.1 GAKP01007235 JAC51717.1 KA646845 AFP61474.1 KQ435710 KOX79756.1 AXCM01000475 CP012526 ALC45833.1 KQ414657 KOC65715.1 CH933806 EDW14278.1 CH477455 EAT40639.1 GL888233 EGI64390.1 CH902617 EDV44252.1 AJVK01028559 GEDC01009110 JAS28188.1 GAMC01006078 JAC00478.1 CH479179 EDW24410.1 CM000070 EAL27967.1 CM000160 EDW98985.1 CH954182 EDV53896.1 AE014297 BT099549 AAF56253.1 ACU32633.1 CM000364 EDX14182.1 CH480815 EDW43452.1 AJWK01016549 AJWK01016550 ATLV01026348 KE525410 KFB53069.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000002358
+ More
UP000215335 UP000019118 UP000030742 UP000007266 UP000000311 UP000078542 UP000075809 UP000008237 UP000027135 UP000053097 UP000279307 UP000005205 UP000037069 UP000235965 UP000078540 UP000078541 UP000095300 UP000095301 UP000078492 UP000007798 UP000091820 UP000001070 UP000092443 UP000092460 UP000075881 UP000075884 UP000076502 UP000005203 UP000069940 UP000249989 UP000075886 UP000092444 UP000092445 UP000078200 UP000075902 UP000075882 UP000007062 UP000075840 UP000075903 UP000076407 UP000069272 UP000000673 UP000007819 UP000076408 UP000075900 UP000008792 UP000192223 UP000075901 UP000053105 UP000075885 UP000075883 UP000092553 UP000053825 UP000075920 UP000075880 UP000009192 UP000008820 UP000007755 UP000007801 UP000092462 UP000008744 UP000001819 UP000002282 UP000008711 UP000000803 UP000000304 UP000001292 UP000092461 UP000030765
UP000215335 UP000019118 UP000030742 UP000007266 UP000000311 UP000078542 UP000075809 UP000008237 UP000027135 UP000053097 UP000279307 UP000005205 UP000037069 UP000235965 UP000078540 UP000078541 UP000095300 UP000095301 UP000078492 UP000007798 UP000091820 UP000001070 UP000092443 UP000092460 UP000075881 UP000075884 UP000076502 UP000005203 UP000069940 UP000249989 UP000075886 UP000092444 UP000092445 UP000078200 UP000075902 UP000075882 UP000007062 UP000075840 UP000075903 UP000076407 UP000069272 UP000000673 UP000007819 UP000076408 UP000075900 UP000008792 UP000192223 UP000075901 UP000053105 UP000075885 UP000075883 UP000092553 UP000053825 UP000075920 UP000075880 UP000009192 UP000008820 UP000007755 UP000007801 UP000092462 UP000008744 UP000001819 UP000002282 UP000008711 UP000000803 UP000000304 UP000001292 UP000092461 UP000030765
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
ProteinModelPortal
H9IZ07
A0A3S2LCY7
A0A1E1W2J7
A0A194QD36
A0A2H1WP44
A0A194RNM0
+ More
A0A212EN86 S4PA54 A0A1Y1MMN2 K7J2C8 A0A232FBB7 A0A0C9QKD3 J3JYQ6 A0A0T6BFK8 D6X4V8 A0A0K8TQ58 E2AB33 E9IHP2 A0A195D463 A0A151WEY8 E2BB57 A0A067QVT5 A0A026WTQ7 A0A158NW01 A0A0L0BN32 A0A2J7QK69 A0A195BN19 A0A195FYC5 A0A1I8NTN3 A0A336MZ64 T1PM15 A0A151J3N9 B4NAG7 A0A1A9WEE5 A0A1B6L7S6 A0A2S2QWP0 A0A1B6II37 B4JH42 A0A1A9XL80 A0A1B0B8E0 A0A182K0K9 A0A182N6K3 A0A154PKV0 A0A088AFK5 A0A1L8DCU4 A0A182GU83 A0A182QLP4 A0A1L8DCE8 A0A1B0GAF7 A0A1A9ZUP8 A0A1A9VGG4 A0A1B6G4M2 A0A023EKW0 A0A182TYR1 A0A182LC08 Q7QBP1 A0A182HIW6 A0A182HD58 A0A182USQ4 A0A182X8B1 A0A2J7QK63 A0A182FPK0 A0A2M4BYE7 W5JA47 A0A2M4BY75 A0A2M3Z8I9 J9JMP3 A0A182XVR5 C4WTU9 A0A182RXZ8 B4MBX2 A0A2M4AHU3 A0A2S2NM15 A0A034WBQ2 A0A1W4WE68 A0A182SQ10 T1PFA3 A0A0M9AAE2 A0A182P2Y6 A0A182MU80 A0A0M3QXF4 A0A0L7R4A4 A0A182W0B3 A0A182IVR8 B4K7F4 Q171H7 F4WN22 B3M0K5 A0A1B0GN99 A0A1B6DR73 W8BNT0 B4G3Z9 Q298I8 B4PL00 B3P776 Q9VCB9 B4QSI3 B4HGJ9 A0A1B0CKR5 A0A084WS75
A0A212EN86 S4PA54 A0A1Y1MMN2 K7J2C8 A0A232FBB7 A0A0C9QKD3 J3JYQ6 A0A0T6BFK8 D6X4V8 A0A0K8TQ58 E2AB33 E9IHP2 A0A195D463 A0A151WEY8 E2BB57 A0A067QVT5 A0A026WTQ7 A0A158NW01 A0A0L0BN32 A0A2J7QK69 A0A195BN19 A0A195FYC5 A0A1I8NTN3 A0A336MZ64 T1PM15 A0A151J3N9 B4NAG7 A0A1A9WEE5 A0A1B6L7S6 A0A2S2QWP0 A0A1B6II37 B4JH42 A0A1A9XL80 A0A1B0B8E0 A0A182K0K9 A0A182N6K3 A0A154PKV0 A0A088AFK5 A0A1L8DCU4 A0A182GU83 A0A182QLP4 A0A1L8DCE8 A0A1B0GAF7 A0A1A9ZUP8 A0A1A9VGG4 A0A1B6G4M2 A0A023EKW0 A0A182TYR1 A0A182LC08 Q7QBP1 A0A182HIW6 A0A182HD58 A0A182USQ4 A0A182X8B1 A0A2J7QK63 A0A182FPK0 A0A2M4BYE7 W5JA47 A0A2M4BY75 A0A2M3Z8I9 J9JMP3 A0A182XVR5 C4WTU9 A0A182RXZ8 B4MBX2 A0A2M4AHU3 A0A2S2NM15 A0A034WBQ2 A0A1W4WE68 A0A182SQ10 T1PFA3 A0A0M9AAE2 A0A182P2Y6 A0A182MU80 A0A0M3QXF4 A0A0L7R4A4 A0A182W0B3 A0A182IVR8 B4K7F4 Q171H7 F4WN22 B3M0K5 A0A1B0GN99 A0A1B6DR73 W8BNT0 B4G3Z9 Q298I8 B4PL00 B3P776 Q9VCB9 B4QSI3 B4HGJ9 A0A1B0CKR5 A0A084WS75
PDB
2EBM
E-value=3.90986e-18,
Score=222
Ontologies
GO
Topology
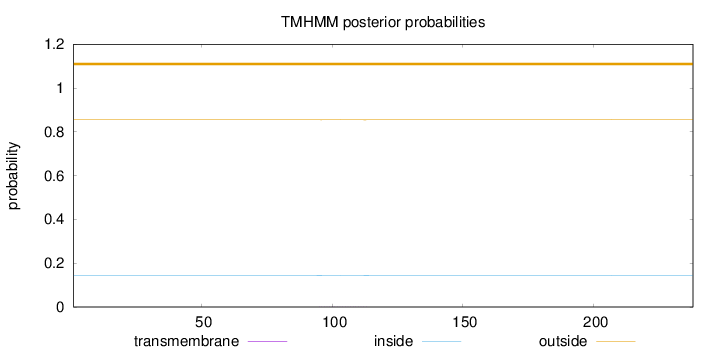
Length:
238
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01794
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.14472
outside
1 - 238
Population Genetic Test Statistics
Pi
193.86581
Theta
187.971766
Tajima's D
0.137578
CLR
0.018255
CSRT
0.405879706014699
Interpretation
Uncertain
