Pre Gene Modal
BGIBMGA002422
Annotation
BRISC_and_BRCA1-A_complex_member_1_[Operophtera_brumata]
Full name
BRISC and BRCA1-A complex member 1
Alternative Name
Mediator of RAP80 interactions and targeting subunit of 40 kDa
New component of the BRCA1-A complex
New component of the BRCA1-A complex
Location in the cell
Nuclear Reliability : 3.295
Sequence
CDS
ATGGAAAGTCCTGAGACAGTATCTTCTAGTAACTCACATGGAGCATCAGACAGTGATTCAAGACAATATGAAGATACGTCTGTCCTCGAGAATGAAGATGTTAAGGAAATACAAGAACAAGTTCTTGCAAATCTAGAGAAGCCTAATCTGCCAAATGTAAATGTACCAGAGAAAATAATTGTCTGCTTAGATGTTTGCTATGACAATCCAAATTCTCTATACCGTTTTGGTGATGGCACAACATTTTCTCCAATCAATATGTTGAAACGAATACTTGATTTCTTTTTTCATACAAAACATGCAATCAGCAAGAAAACTGAATTTGCATTGATAACACTTCAAGATGCAGGTGCATGTTGGACCCAGAATTTTACAAGCAATGTTAAAGATTTAATCAGTGCTATTGATTATGCACATGCTGAAGAAGCTACATCAGACACTTTTGATTTTGAAAAAGTATATGAACTAATAAGACATCATATAGAAATTCCTATGATGAAACCTGGAGAATCCTTACTGCCTCCTCCTTATATCGTCAGACTTCTGATTTTATATGGAAGATCTAATTGTGTTCCAGTAATTCCTCATGAAGACCCATGTTTCCAATATATAAGAAGTCAGATATATTTTTATACAGATATTCTGTTGGCACATGAAGATGATTGTGCTACGCATAAATGTGATGAAATATATGATGCATTACAGGATCTTGATAATGGCTATTCTTATGTGTTTGAGGTATCAAGAAATGCTACTAAAATACATGATTGTATTGCAAAATTGTTAGCACATCCACTACAGAGGCCCTTGCAAAAAAATACAGACTATTCTTTCGGTTCAAGATACTAG
Protein
MESPETVSSSNSHGASDSDSRQYEDTSVLENEDVKEIQEQVLANLEKPNLPNVNVPEKIIVCLDVCYDNPNSLYRFGDGTTFSPINMLKRILDFFFHTKHAISKKTEFALITLQDAGACWTQNFTSNVKDLISAIDYAHAEEATSDTFDFEKVYELIRHHIEIPMMKPGESLLPPPYIVRLLILYGRSNCVPVIPHEDPCFQYIRSQIYFYTDILLAHEDDCATHKCDEIYDALQDLDNGYSYVFEVSRNATKIHDCIAKLLAHPLQRPLQKNTDYSFGSRY
Summary
Description
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it is required for the complex integrity and its localization at DSBs. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates. In these 2 complexes, it is probably required to maintain the stability of BABAM2 and help the 'Lys-63'-linked deubiquitinase activity mediated by BRCC3/BRCC36 component. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1. Plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression. Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination.
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it is required for the complex integrity and its localization at DSBs. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates (PubMed:24075985, PubMed:26195665). In these 2 complexes, it is probably required to maintain the stability of BABAM2 and help the 'Lys-63'-linked deubiquitinase activity mediated by BRCC3/BRCC36 component. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). Plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression (PubMed:24075985). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985).
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it is required for the complex integrity and its localization at DSBs. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates (PubMed:24075985, PubMed:26195665). In these 2 complexes, it is probably required to maintain the stability of BABAM2 and help the 'Lys-63'-linked deubiquitinase activity mediated by BRCC3/BRCC36 component. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). Plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression (PubMed:24075985). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985).
Subunit
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1 and BABAM2. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Identified in a complex with SHMT2 and the other subunits of the BRISC complex.
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:24075985). Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19261746, PubMed:19261749, PubMed:19261748, PubMed:21282113). In the BRCA1-A complex, interacts directly with ABRAXAS1 and BABAM2 (PubMed:19261749, PubMed:19261748). Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19214193, PubMed:21282113, PubMed:24075985, PubMed:25283148). Identified in a complex with SHMT2 and the other subunits of the BRISC complex (PubMed:24075985).
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:24075985). Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19261746, PubMed:19261749, PubMed:19261748, PubMed:21282113). In the BRCA1-A complex, interacts directly with ABRAXAS1 and BABAM2 (PubMed:19261749, PubMed:19261748). Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19214193, PubMed:21282113, PubMed:24075985, PubMed:25283148). Identified in a complex with SHMT2 and the other subunits of the BRISC complex (PubMed:24075985).
Similarity
Belongs to the BABAM1 family.
Keywords
Acetylation
Cell cycle
Cell division
Chromatin regulator
Complete proteome
Cytoplasm
DNA damage
DNA repair
Mitosis
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation pathway
Alternative splicing
Feature
chain BRISC and BRCA1-A complex member 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A1E1W822
A0A0L7KZS2
A0A194RMF5
A0A2H1WQY7
A0A212ENB2
A0A194QBU4
+ More
S4PY62 A0A2J7R1Z5 A0A2Z5TTW4 A0A067R4I8 A0A2P8Z3V2 A0A224YHV2 A0A131YPU8 A0A293M7V1 A0A210PP11 A0A0J7KGL3 D6WBJ6 W4XNB0 A0A1S3IKZ1 A0A1E1X5U3 E2ACB1 A0A131XM43 L7M6Y1 A0A2C9KMD8 A0A2R5LDX1 A0A2L2YBN6 E2BAT3 A0A310SKB5 A0A195FFX0 A0A1W4X5A3 A0A151XCY5 F4WZP3 A0A195B7L7 A0A158NGT6 E9IUQ5 A0A195DYB1 N6TFH4 U4ULC8 A0A131XSE7 J3JYK6 A0A026WYT5 A0A1Y1LX77 A0A0B7A8H0 A0A0B7AAM2 A0A3M6UGC0 A0A0C9RUJ1 A0A2T7NNE0 A0A088AN10 A7SJ60 B7PWX5 A0A2L2Y9D3 A0A2A3EE98 V9KVI4 H3AMY9 A0A0L8H8J2 A0A2G8KE64 Q5XIJ6 A0A1U8D1I6 A0A337RVM9 A0A0S7H745 A0A3Q1AHR9 A0A3Q2R1H1 H0WYE5 A0A3Q3GXI1 A0A2K5DJY1 A0A3B3S731 Q3UI43 A0A024R7L2 Q9NWV8 A0A2Y9GI90 A0A2J8T6F8 Q5R7L2 G3S649 K7CLA9 A0A2K5SDJ2 A0A286ZQV9 A0A3Q4I3H0 A0A3Q7Q675 A0A2U3ZJX1 A0A384D5D7 G1LV83 A0A3Q7W4Q4 A0A2K6UFN5 A0A3B3VAS9 E2R851 A0A3Q7RTP5 I3JV16 A0A1U7T970 A0A3P9AF93 M3ZJJ1 A0A2U3Z1D6 A0A2K6MYK1 A0A2K6PVV9 A0A2K5LT51 A0A0D9R019 A0A2I3M2C4 A0A1U7RRD5
S4PY62 A0A2J7R1Z5 A0A2Z5TTW4 A0A067R4I8 A0A2P8Z3V2 A0A224YHV2 A0A131YPU8 A0A293M7V1 A0A210PP11 A0A0J7KGL3 D6WBJ6 W4XNB0 A0A1S3IKZ1 A0A1E1X5U3 E2ACB1 A0A131XM43 L7M6Y1 A0A2C9KMD8 A0A2R5LDX1 A0A2L2YBN6 E2BAT3 A0A310SKB5 A0A195FFX0 A0A1W4X5A3 A0A151XCY5 F4WZP3 A0A195B7L7 A0A158NGT6 E9IUQ5 A0A195DYB1 N6TFH4 U4ULC8 A0A131XSE7 J3JYK6 A0A026WYT5 A0A1Y1LX77 A0A0B7A8H0 A0A0B7AAM2 A0A3M6UGC0 A0A0C9RUJ1 A0A2T7NNE0 A0A088AN10 A7SJ60 B7PWX5 A0A2L2Y9D3 A0A2A3EE98 V9KVI4 H3AMY9 A0A0L8H8J2 A0A2G8KE64 Q5XIJ6 A0A1U8D1I6 A0A337RVM9 A0A0S7H745 A0A3Q1AHR9 A0A3Q2R1H1 H0WYE5 A0A3Q3GXI1 A0A2K5DJY1 A0A3B3S731 Q3UI43 A0A024R7L2 Q9NWV8 A0A2Y9GI90 A0A2J8T6F8 Q5R7L2 G3S649 K7CLA9 A0A2K5SDJ2 A0A286ZQV9 A0A3Q4I3H0 A0A3Q7Q675 A0A2U3ZJX1 A0A384D5D7 G1LV83 A0A3Q7W4Q4 A0A2K6UFN5 A0A3B3VAS9 E2R851 A0A3Q7RTP5 I3JV16 A0A1U7T970 A0A3P9AF93 M3ZJJ1 A0A2U3Z1D6 A0A2K6MYK1 A0A2K6PVV9 A0A2K5LT51 A0A0D9R019 A0A2I3M2C4 A0A1U7RRD5
Pubmed
26227816
26354079
22118469
23622113
26760975
24845553
+ More
29403074 28797301 26830274 28812685 18362917 19820115 28503490 20798317 28049606 25576852 15562597 26561354 21719571 21347285 21282665 23537049 29652888 22516182 24508170 30249741 28004739 30382153 17615350 24402279 9215903 29023486 15489334 22673903 28071753 17975172 29240929 16141072 21183079 11181995 11042152 11230166 14702039 15057824 17081983 18691976 18669648 19413330 19214193 19261746 19261749 19261748 19690332 20068231 21269460 21282113 21406692 24075985 23186163 24275569 25283148 26195665 22398555 16136131 25186727 24813606 20010809 16341006 25069045 23542700 25362486
29403074 28797301 26830274 28812685 18362917 19820115 28503490 20798317 28049606 25576852 15562597 26561354 21719571 21347285 21282665 23537049 29652888 22516182 24508170 30249741 28004739 30382153 17615350 24402279 9215903 29023486 15489334 22673903 28071753 17975172 29240929 16141072 21183079 11181995 11042152 11230166 14702039 15057824 17081983 18691976 18669648 19413330 19214193 19261746 19261749 19261748 19690332 20068231 21269460 21282113 21406692 24075985 23186163 24275569 25283148 26195665 22398555 16136131 25186727 24813606 20010809 16341006 25069045 23542700 25362486
EMBL
GDQN01007959
JAT83095.1
JTDY01004119
KOB68576.1
KQ459984
KPJ19008.1
+ More
ODYU01010028 SOQ54844.1 AGBW02013684 OWR42966.1 KQ459220 KPJ02889.1 GAIX01003843 JAA88717.1 NEVH01008202 PNF34864.1 FX985757 BBA93644.1 KK852700 KDR18145.1 PYGN01000208 PSN51169.1 GFPF01002637 MAA13783.1 GEDV01007273 JAP81284.1 GFWV01011540 MAA36269.1 NEDP02005572 OWF38176.1 LBMM01007833 KMQ89384.1 KQ971311 EEZ98909.1 AAGJ04093207 GFAC01004541 JAT94647.1 GL438491 EFN68901.1 GEFH01001383 JAP67198.1 GACK01006160 JAA58874.1 GGLE01003489 MBY07615.1 IAAA01016470 LAA04780.1 GL446831 EFN87218.1 KQ761768 OAD57126.1 KQ981625 KYN39122.1 KQ982294 KYQ58209.1 GL888480 EGI60318.1 KQ976574 KYM80240.1 ADTU01015209 GL766012 EFZ15701.1 KQ980107 KYN17692.1 APGK01029756 KB740741 ENN79104.1 KB632375 ERL93912.1 GEFM01005592 GEGO01002018 JAP70204.1 JAR93386.1 BT128335 AEE63293.1 KK107063 QOIP01000005 EZA61177.1 RLU22354.1 GEZM01050287 JAV75637.1 HACG01030203 CEK77068.1 HACG01030201 CEK77066.1 RCHS01001584 RMX52731.1 GBYB01012519 GBYB01012520 JAG82286.1 JAG82287.1 PZQS01000010 PVD22690.1 DS469675 EDO36240.1 ABJB010994087 ABJB011119944 DS810815 EEC11097.1 IAAA01016471 LAA04782.1 KZ288269 PBC30038.1 JW869780 AFP02298.1 AFYH01122492 AFYH01122493 KQ418900 KOF85414.1 MRZV01000653 PIK46294.1 BC083685 AANG04000016 GBYX01445955 JAO35485.1 AAQR03128734 AK003366 AK077402 AK078265 AK147083 BC005692 CH471106 EAW84585.1 AF161491 AL136692 AK000578 AK299493 AK301193 CR533526 AC104522 BC000788 BC006244 BC091491 NDHI03003519 PNJ28625.1 PNJ28626.1 PNJ28627.1 CR860103 CABD030112167 CABD030112168 CABD030112169 AACZ04031524 GABC01007207 GABC01007206 GABF01000030 GABF01000029 GABD01008105 GABD01008104 NBAG03000278 JAA04131.1 JAA04132.1 JAA22115.1 JAA24995.1 PNI50674.1 PNI50675.1 PNI50676.1 AEMK02000010 ACTA01096567 AAEX03012297 AERX01013022 AQIB01141369 AQIB01141370 AHZZ02013755
ODYU01010028 SOQ54844.1 AGBW02013684 OWR42966.1 KQ459220 KPJ02889.1 GAIX01003843 JAA88717.1 NEVH01008202 PNF34864.1 FX985757 BBA93644.1 KK852700 KDR18145.1 PYGN01000208 PSN51169.1 GFPF01002637 MAA13783.1 GEDV01007273 JAP81284.1 GFWV01011540 MAA36269.1 NEDP02005572 OWF38176.1 LBMM01007833 KMQ89384.1 KQ971311 EEZ98909.1 AAGJ04093207 GFAC01004541 JAT94647.1 GL438491 EFN68901.1 GEFH01001383 JAP67198.1 GACK01006160 JAA58874.1 GGLE01003489 MBY07615.1 IAAA01016470 LAA04780.1 GL446831 EFN87218.1 KQ761768 OAD57126.1 KQ981625 KYN39122.1 KQ982294 KYQ58209.1 GL888480 EGI60318.1 KQ976574 KYM80240.1 ADTU01015209 GL766012 EFZ15701.1 KQ980107 KYN17692.1 APGK01029756 KB740741 ENN79104.1 KB632375 ERL93912.1 GEFM01005592 GEGO01002018 JAP70204.1 JAR93386.1 BT128335 AEE63293.1 KK107063 QOIP01000005 EZA61177.1 RLU22354.1 GEZM01050287 JAV75637.1 HACG01030203 CEK77068.1 HACG01030201 CEK77066.1 RCHS01001584 RMX52731.1 GBYB01012519 GBYB01012520 JAG82286.1 JAG82287.1 PZQS01000010 PVD22690.1 DS469675 EDO36240.1 ABJB010994087 ABJB011119944 DS810815 EEC11097.1 IAAA01016471 LAA04782.1 KZ288269 PBC30038.1 JW869780 AFP02298.1 AFYH01122492 AFYH01122493 KQ418900 KOF85414.1 MRZV01000653 PIK46294.1 BC083685 AANG04000016 GBYX01445955 JAO35485.1 AAQR03128734 AK003366 AK077402 AK078265 AK147083 BC005692 CH471106 EAW84585.1 AF161491 AL136692 AK000578 AK299493 AK301193 CR533526 AC104522 BC000788 BC006244 BC091491 NDHI03003519 PNJ28625.1 PNJ28626.1 PNJ28627.1 CR860103 CABD030112167 CABD030112168 CABD030112169 AACZ04031524 GABC01007207 GABC01007206 GABF01000030 GABF01000029 GABD01008105 GABD01008104 NBAG03000278 JAA04131.1 JAA04132.1 JAA22115.1 JAA24995.1 PNI50674.1 PNI50675.1 PNI50676.1 AEMK02000010 ACTA01096567 AAEX03012297 AERX01013022 AQIB01141369 AQIB01141370 AHZZ02013755
Proteomes
UP000037510
UP000053240
UP000007151
UP000053268
UP000235965
UP000027135
+ More
UP000245037 UP000242188 UP000036403 UP000007266 UP000007110 UP000085678 UP000000311 UP000076420 UP000008237 UP000078541 UP000192223 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000019118 UP000030742 UP000053097 UP000279307 UP000275408 UP000245119 UP000005203 UP000001593 UP000001555 UP000242457 UP000008672 UP000053454 UP000230750 UP000002494 UP000189706 UP000011712 UP000257160 UP000265000 UP000005225 UP000261660 UP000233020 UP000261540 UP000000589 UP000005640 UP000248481 UP000001595 UP000001519 UP000002277 UP000233040 UP000008227 UP000261580 UP000286641 UP000245340 UP000261680 UP000291021 UP000008912 UP000286642 UP000233220 UP000261500 UP000002254 UP000286640 UP000005207 UP000189704 UP000265140 UP000002852 UP000245341 UP000233180 UP000233200 UP000233060 UP000029965 UP000028761 UP000189705
UP000245037 UP000242188 UP000036403 UP000007266 UP000007110 UP000085678 UP000000311 UP000076420 UP000008237 UP000078541 UP000192223 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000019118 UP000030742 UP000053097 UP000279307 UP000275408 UP000245119 UP000005203 UP000001593 UP000001555 UP000242457 UP000008672 UP000053454 UP000230750 UP000002494 UP000189706 UP000011712 UP000257160 UP000265000 UP000005225 UP000261660 UP000233020 UP000261540 UP000000589 UP000005640 UP000248481 UP000001595 UP000001519 UP000002277 UP000233040 UP000008227 UP000261580 UP000286641 UP000245340 UP000261680 UP000291021 UP000008912 UP000286642 UP000233220 UP000261500 UP000002254 UP000286640 UP000005207 UP000189704 UP000265140 UP000002852 UP000245341 UP000233180 UP000233200 UP000233060 UP000029965 UP000028761 UP000189705
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
A0A1E1W822
A0A0L7KZS2
A0A194RMF5
A0A2H1WQY7
A0A212ENB2
A0A194QBU4
+ More
S4PY62 A0A2J7R1Z5 A0A2Z5TTW4 A0A067R4I8 A0A2P8Z3V2 A0A224YHV2 A0A131YPU8 A0A293M7V1 A0A210PP11 A0A0J7KGL3 D6WBJ6 W4XNB0 A0A1S3IKZ1 A0A1E1X5U3 E2ACB1 A0A131XM43 L7M6Y1 A0A2C9KMD8 A0A2R5LDX1 A0A2L2YBN6 E2BAT3 A0A310SKB5 A0A195FFX0 A0A1W4X5A3 A0A151XCY5 F4WZP3 A0A195B7L7 A0A158NGT6 E9IUQ5 A0A195DYB1 N6TFH4 U4ULC8 A0A131XSE7 J3JYK6 A0A026WYT5 A0A1Y1LX77 A0A0B7A8H0 A0A0B7AAM2 A0A3M6UGC0 A0A0C9RUJ1 A0A2T7NNE0 A0A088AN10 A7SJ60 B7PWX5 A0A2L2Y9D3 A0A2A3EE98 V9KVI4 H3AMY9 A0A0L8H8J2 A0A2G8KE64 Q5XIJ6 A0A1U8D1I6 A0A337RVM9 A0A0S7H745 A0A3Q1AHR9 A0A3Q2R1H1 H0WYE5 A0A3Q3GXI1 A0A2K5DJY1 A0A3B3S731 Q3UI43 A0A024R7L2 Q9NWV8 A0A2Y9GI90 A0A2J8T6F8 Q5R7L2 G3S649 K7CLA9 A0A2K5SDJ2 A0A286ZQV9 A0A3Q4I3H0 A0A3Q7Q675 A0A2U3ZJX1 A0A384D5D7 G1LV83 A0A3Q7W4Q4 A0A2K6UFN5 A0A3B3VAS9 E2R851 A0A3Q7RTP5 I3JV16 A0A1U7T970 A0A3P9AF93 M3ZJJ1 A0A2U3Z1D6 A0A2K6MYK1 A0A2K6PVV9 A0A2K5LT51 A0A0D9R019 A0A2I3M2C4 A0A1U7RRD5
S4PY62 A0A2J7R1Z5 A0A2Z5TTW4 A0A067R4I8 A0A2P8Z3V2 A0A224YHV2 A0A131YPU8 A0A293M7V1 A0A210PP11 A0A0J7KGL3 D6WBJ6 W4XNB0 A0A1S3IKZ1 A0A1E1X5U3 E2ACB1 A0A131XM43 L7M6Y1 A0A2C9KMD8 A0A2R5LDX1 A0A2L2YBN6 E2BAT3 A0A310SKB5 A0A195FFX0 A0A1W4X5A3 A0A151XCY5 F4WZP3 A0A195B7L7 A0A158NGT6 E9IUQ5 A0A195DYB1 N6TFH4 U4ULC8 A0A131XSE7 J3JYK6 A0A026WYT5 A0A1Y1LX77 A0A0B7A8H0 A0A0B7AAM2 A0A3M6UGC0 A0A0C9RUJ1 A0A2T7NNE0 A0A088AN10 A7SJ60 B7PWX5 A0A2L2Y9D3 A0A2A3EE98 V9KVI4 H3AMY9 A0A0L8H8J2 A0A2G8KE64 Q5XIJ6 A0A1U8D1I6 A0A337RVM9 A0A0S7H745 A0A3Q1AHR9 A0A3Q2R1H1 H0WYE5 A0A3Q3GXI1 A0A2K5DJY1 A0A3B3S731 Q3UI43 A0A024R7L2 Q9NWV8 A0A2Y9GI90 A0A2J8T6F8 Q5R7L2 G3S649 K7CLA9 A0A2K5SDJ2 A0A286ZQV9 A0A3Q4I3H0 A0A3Q7Q675 A0A2U3ZJX1 A0A384D5D7 G1LV83 A0A3Q7W4Q4 A0A2K6UFN5 A0A3B3VAS9 E2R851 A0A3Q7RTP5 I3JV16 A0A1U7T970 A0A3P9AF93 M3ZJJ1 A0A2U3Z1D6 A0A2K6MYK1 A0A2K6PVV9 A0A2K5LT51 A0A0D9R019 A0A2I3M2C4 A0A1U7RRD5
Ontologies
KEGG
GO
PANTHER
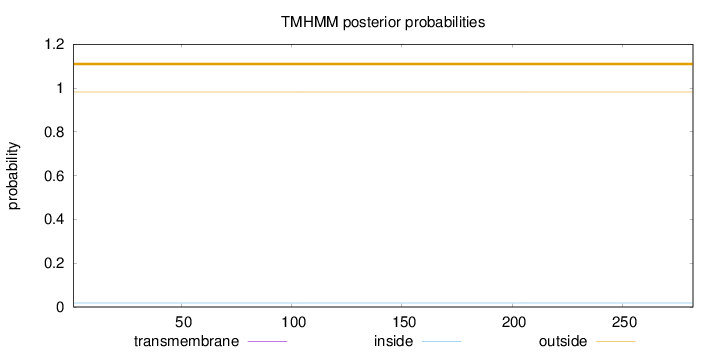
Topology
Subcellular location
Cytoplasm
Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 2 publications.
Nucleus Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 2 publications.
Nucleus Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 2 publications.
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00169
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01798
outside
1 - 282
Population Genetic Test Statistics
Pi
206.789372
Theta
174.080653
Tajima's D
0.586308
CLR
0
CSRT
0.540272986350682
Interpretation
Uncertain
