Pre Gene Modal
BGIBMGA002424
Annotation
PREDICTED:_STAM-binding_protein-like_A_[Amyelois_transitella]
Full name
STAM-binding protein-like A
Location in the cell
Nuclear Reliability : 2.727
Sequence
CDS
ATGCAGTTGGAAGAAAAACGCCCCAAACATGTTGATCTAGCTTCAATGGAACCTTCAGCAAGAGTTAAGCAGCTCGCAAACTACGGGACGATGGTTGACGTCGATCCTAATGTTCCACCTAGAAGGTACTATAGATCTGGTCTTGAGATGGTAAGAATGGCTAATGTGTATCTAGCAGAAGGGAGCTTGGAGAATGCATATATTCTATATATGAAGTTCATGACACTATTCCTGGAAAAGATACGGAAACATCCAGAATACAGTAGTGTGCCTGCACAAGTGAAATCTGTCAATCAGGGTAAACTCAAAGAGGTGATGCCTAAAGCTGAGAAACTGAAACAAAAGCTCTTGGATCAGTATGCTAAAGAACATGCCCAGTATAAAGAGAATGAGGAAAAGCGACAGATTGCCGAAGCACAAAAGAAGAAACAAGAGCAGGCCGATGCGAAACTCGCTGAAAGATTACAAGCAGATGAAAATAAGAGAGATGGAAACTCAACAATCCCTCACTTATTGAATAATGAACAATGGGTAGTTTCACCGTCTGCACCAGCTGTTGATAGGGTCTTGTATCCTGATGACTTTGCAGCAGAACCTCCGCAAGCACCTCCCCATCACTACCAACCTGTGCCACCTCTTATACCACCTTCAAGGCCACATAATGACAGCAGTCTTGGGGGTGCGTTGTACGGTTCTCACCGACTCCGCACCGTGGTTGTGCCCGCGGCGTTACTCGGACGATTCTTGCGTCTCGCTGCCGATAATACCACACGAAATATAGAGACCTGTGGTATACTTGCGGGGATACTGGAGCGGAACCAGTTGAAGATCACACACGTTATAGTCCCAAAGCAGTCCGGTACCCCGGACTCATGCAGCACCAACAACGAAGAGGAGATATTTCACTACCAGGACCAACATAACCTCATCACACTAGGATGGATACATACGCATCCGACGCAGACCGCGTTCCTGTCGTCGGTGGATCTGCACACGCAGTGCTCGTACCAGCTGATGATGGCCGAAGCGATCGCGATCGTTTGCGCGCCCAAATACAACGAAACTGGCTACTTCGCCTTAACACCGGACTACGGTATGCAGTTCATAGCGAACTGTAGACAGAGCGGCTTCCACCCGCACCCCAATGACCCGCCTCTGTTTCACAATGTATCACATATAAACGTAGACGACGCAGCTCCTATAGAAATGGTCGATCTCAGGAGATGA
Protein
MQLEEKRPKHVDLASMEPSARVKQLANYGTMVDVDPNVPPRRYYRSGLEMVRMANVYLAEGSLENAYILYMKFMTLFLEKIRKHPEYSSVPAQVKSVNQGKLKEVMPKAEKLKQKLLDQYAKEHAQYKENEEKRQIAEAQKKKQEQADAKLAERLQADENKRDGNSTIPHLLNNEQWVVSPSAPAVDRVLYPDDFAAEPPQAPPHHYQPVPPLIPPSRPHNDSSLGGALYGSHRLRTVVVPAALLGRFLRLAADNTTRNIETCGILAGILERNQLKITHVIVPKQSGTPDSCSTNNEEEIFHYQDQHNLITLGWIHTHPTQTAFLSSVDLHTQCSYQLMMAEAIAIVCAPKYNETGYFALTPDYGMQFIANCRQSGFHPHPNDPPLFHNVSHINVDDAAPIEMVDLRR
Summary
Description
Zinc metalloprotease that specifically cleaves 'Lys-63'-linked polyubiquitin chains. Does not cleave 'Lys-48'-linked polyubiquitin chains (By similarity). Functions at the endosome and is able to oppose the ubiquitin-dependent sorting of receptors to lysosomes (By similarity).
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M67C family.
Keywords
Complete proteome
Hydrolase
Metal-binding
Metalloprotease
Protease
Reference proteome
Ubl conjugation pathway
Zinc
Feature
chain STAM-binding protein-like A
Uniprot
H9IYT9
A0A2A4JEZ5
A0A194QBG4
A0A212EHR8
A0A2H1WJZ4
A0A2A4IT09
+ More
A0A336MJ09 A0A336L450 A0A067RMR1 A0A1L8DME8 A0A0K8TKB2 D6X4C6 A0A1Y1JXA6 B0W7H5 A0A1Q3F798 N6UE47 B4JVA3 U4U056 A0A1I8PTP5 W8BLD5 A0A154PU72 B4LYM2 A0A034VZX0 A0A0A9XYL0 A0A0K8UQY4 A0A0A1X9R0 Q9VA71 B4R1M5 Q8T8R5 B4PLW8 A0A3L8DD54 B3P585 B4HZR2 B4NGW0 A0A0L0BLE6 A0A1A9XTD2 B4K8R5 E0VIA9 A0A1B0BUR2 Q176G0 A0A1I8MC82 A0A3B0K3A1 A0A1W4W458 Q29C42 A0A1A9ZZK4 A0A0M3QYH5 A0A151IQG2 A0A158NU93 A0A1B0FR88 B3MTC0 A0A0P5ECE0 F4WR70 A0A151JCT4 A0A1A9WN38 A0A1B6DP92 A0A195BBN9 A0A0N0BF55 A0A0P4WA17 A0A232FC62 K7J2B7 E9H3M7 R7UEM3 A0A0P5YAE6 A0A1B6G315 C3XZW8 A0A0P6IFD4 A0A3B3YK73 A0A1Z5L619 A0A1B6HV73 A0A293LZQ2 A0A3B3QTS4 A0A2B4RMA9 A0A3B4EER2 E2BU25 Q6TH47 A0A2D0QZI6 F1QBG8 A0A2R5LNF0 A0A3B3X5Z8 U3ID15 A0A3M6V5R6 A0A087XLE7 A0A1A9VVI2 A0A1S3PKR0 R0L8X6 K7FH03 A0A3B3UUW0 A0A0L8GQP8 A0A3N0Z715 A0A0L8GRS4 M4AM50 F7EMR4 W5KDA0 A0A3P8Y729 K7FH21 A0A1A7X4E7 A0A1U7RXZ4 V9KWF6 R7VTM2 A0A093J235
A0A336MJ09 A0A336L450 A0A067RMR1 A0A1L8DME8 A0A0K8TKB2 D6X4C6 A0A1Y1JXA6 B0W7H5 A0A1Q3F798 N6UE47 B4JVA3 U4U056 A0A1I8PTP5 W8BLD5 A0A154PU72 B4LYM2 A0A034VZX0 A0A0A9XYL0 A0A0K8UQY4 A0A0A1X9R0 Q9VA71 B4R1M5 Q8T8R5 B4PLW8 A0A3L8DD54 B3P585 B4HZR2 B4NGW0 A0A0L0BLE6 A0A1A9XTD2 B4K8R5 E0VIA9 A0A1B0BUR2 Q176G0 A0A1I8MC82 A0A3B0K3A1 A0A1W4W458 Q29C42 A0A1A9ZZK4 A0A0M3QYH5 A0A151IQG2 A0A158NU93 A0A1B0FR88 B3MTC0 A0A0P5ECE0 F4WR70 A0A151JCT4 A0A1A9WN38 A0A1B6DP92 A0A195BBN9 A0A0N0BF55 A0A0P4WA17 A0A232FC62 K7J2B7 E9H3M7 R7UEM3 A0A0P5YAE6 A0A1B6G315 C3XZW8 A0A0P6IFD4 A0A3B3YK73 A0A1Z5L619 A0A1B6HV73 A0A293LZQ2 A0A3B3QTS4 A0A2B4RMA9 A0A3B4EER2 E2BU25 Q6TH47 A0A2D0QZI6 F1QBG8 A0A2R5LNF0 A0A3B3X5Z8 U3ID15 A0A3M6V5R6 A0A087XLE7 A0A1A9VVI2 A0A1S3PKR0 R0L8X6 K7FH03 A0A3B3UUW0 A0A0L8GQP8 A0A3N0Z715 A0A0L8GRS4 M4AM50 F7EMR4 W5KDA0 A0A3P8Y729 K7FH21 A0A1A7X4E7 A0A1U7RXZ4 V9KWF6 R7VTM2 A0A093J235
EC Number
3.4.19.-
Pubmed
19121390
26354079
22118469
24845553
26369729
18362917
+ More
19820115 28004739 23537049 17994087 24495485 25348373 25401762 26823975 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26108605 20566863 17510324 25315136 15632085 21347285 21719571 28648823 20075255 21292972 23254933 18563158 28528879 29240929 20798317 15520368 23594743 23749191 30382153 17381049 23542700 20431018 25329095 25069045 24402279 23371554
19820115 28004739 23537049 17994087 24495485 25348373 25401762 26823975 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26108605 20566863 17510324 25315136 15632085 21347285 21719571 28648823 20075255 21292972 23254933 18563158 28528879 29240929 20798317 15520368 23594743 23749191 30382153 17381049 23542700 20431018 25329095 25069045 24402279 23371554
EMBL
BABH01010846
NWSH01001719
PCG70346.1
KQ459220
KPJ02883.1
AGBW02014807
+ More
OWR41029.1 ODYU01009163 SOQ53403.1 NWSH01007570 PCG62905.1 UFQT01001304 SSX29920.1 UFQS01001467 UFQT01001467 SSX11127.1 SSX30698.1 KK852417 KDR24328.1 GFDF01006462 JAV07622.1 GDAI01002919 JAI14684.1 KQ971410 EEZ97535.1 GEZM01098748 JAV53773.1 DS231854 EDS38010.1 GFDL01011604 JAV23441.1 APGK01026102 KB740615 ENN79985.1 CH916374 EDV91423.1 KB631815 ERL86452.1 GAMC01006948 JAB99607.1 KQ435180 KZC14994.1 CH940650 EDW68042.1 GAKP01010106 JAC48846.1 GBHO01018610 GDHC01009153 JAG24994.1 JAQ09476.1 GDHF01023318 JAI28996.1 GBXI01006253 GBXI01000039 JAD08039.1 JAD14253.1 AE014297 BT133327 AAF57051.1 AFE85380.1 CM000364 EDX15013.1 AY075309 AAL68176.1 CM000160 EDW99105.1 QOIP01000009 RLU18261.1 CH954182 EDV53068.1 CH480819 EDW53519.1 CH964272 EDW84457.1 JRES01001702 KNC20778.1 CH933806 EDW15484.1 DS235184 EEB13115.1 JXJN01020881 JXJN01020882 CH477387 EAT42056.1 OUUW01000005 SPP80096.1 CM000070 EAL26804.1 CP012526 ALC47629.1 KQ976780 KYN08391.1 ADTU01026313 CCAG010022123 CH902623 EDV30510.1 GDIP01143999 GDIQ01081659 LRGB01000568 JAJ79403.1 JAN13078.1 KZS17970.1 GL888284 EGI63297.1 KQ979048 KYN23227.1 GEDC01009800 JAS27498.1 KQ976532 KYM81625.1 KQ435812 KOX72861.1 GDRN01087612 JAI61015.1 NNAY01000456 OXU28265.1 AAZX01001561 GL732589 EFX73539.1 AMQN01001639 KB304239 ELU02238.1 GDIP01062053 JAM41662.1 GECZ01012933 JAS56836.1 GG666477 EEN66334.1 GDIQ01005345 JAN89392.1 GFJQ02004114 JAW02856.1 GECU01029166 JAS78540.1 GFWV01008807 MAA33536.1 LSMT01000470 PFX17477.1 GL450575 EFN80799.1 AY398309 BC055512 BC171484 BX005309 CR925773 GGLE01006822 MBY10948.1 ADON01064568 RCHS01000065 RMX61231.1 AYCK01002589 AYCK01002590 KB742995 EOB02104.1 AGCU01127871 AGCU01127872 KQ420799 KOF79313.1 RJVU01007007 ROL54121.1 KOF79310.1 AAMC01014187 HADW01011284 HADW01014888 HADX01012714 SBP12684.1 JW870395 AFP02913.1 KB375463 AKCR02000216 EMC87227.1 PKK18274.1 KK604060 KFW07973.1
OWR41029.1 ODYU01009163 SOQ53403.1 NWSH01007570 PCG62905.1 UFQT01001304 SSX29920.1 UFQS01001467 UFQT01001467 SSX11127.1 SSX30698.1 KK852417 KDR24328.1 GFDF01006462 JAV07622.1 GDAI01002919 JAI14684.1 KQ971410 EEZ97535.1 GEZM01098748 JAV53773.1 DS231854 EDS38010.1 GFDL01011604 JAV23441.1 APGK01026102 KB740615 ENN79985.1 CH916374 EDV91423.1 KB631815 ERL86452.1 GAMC01006948 JAB99607.1 KQ435180 KZC14994.1 CH940650 EDW68042.1 GAKP01010106 JAC48846.1 GBHO01018610 GDHC01009153 JAG24994.1 JAQ09476.1 GDHF01023318 JAI28996.1 GBXI01006253 GBXI01000039 JAD08039.1 JAD14253.1 AE014297 BT133327 AAF57051.1 AFE85380.1 CM000364 EDX15013.1 AY075309 AAL68176.1 CM000160 EDW99105.1 QOIP01000009 RLU18261.1 CH954182 EDV53068.1 CH480819 EDW53519.1 CH964272 EDW84457.1 JRES01001702 KNC20778.1 CH933806 EDW15484.1 DS235184 EEB13115.1 JXJN01020881 JXJN01020882 CH477387 EAT42056.1 OUUW01000005 SPP80096.1 CM000070 EAL26804.1 CP012526 ALC47629.1 KQ976780 KYN08391.1 ADTU01026313 CCAG010022123 CH902623 EDV30510.1 GDIP01143999 GDIQ01081659 LRGB01000568 JAJ79403.1 JAN13078.1 KZS17970.1 GL888284 EGI63297.1 KQ979048 KYN23227.1 GEDC01009800 JAS27498.1 KQ976532 KYM81625.1 KQ435812 KOX72861.1 GDRN01087612 JAI61015.1 NNAY01000456 OXU28265.1 AAZX01001561 GL732589 EFX73539.1 AMQN01001639 KB304239 ELU02238.1 GDIP01062053 JAM41662.1 GECZ01012933 JAS56836.1 GG666477 EEN66334.1 GDIQ01005345 JAN89392.1 GFJQ02004114 JAW02856.1 GECU01029166 JAS78540.1 GFWV01008807 MAA33536.1 LSMT01000470 PFX17477.1 GL450575 EFN80799.1 AY398309 BC055512 BC171484 BX005309 CR925773 GGLE01006822 MBY10948.1 ADON01064568 RCHS01000065 RMX61231.1 AYCK01002589 AYCK01002590 KB742995 EOB02104.1 AGCU01127871 AGCU01127872 KQ420799 KOF79313.1 RJVU01007007 ROL54121.1 KOF79310.1 AAMC01014187 HADW01011284 HADW01014888 HADX01012714 SBP12684.1 JW870395 AFP02913.1 KB375463 AKCR02000216 EMC87227.1 PKK18274.1 KK604060 KFW07973.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000027135
UP000007266
+ More
UP000002320 UP000019118 UP000001070 UP000030742 UP000095300 UP000076502 UP000008792 UP000000803 UP000000304 UP000002282 UP000279307 UP000008711 UP000001292 UP000007798 UP000037069 UP000092443 UP000009192 UP000009046 UP000092460 UP000008820 UP000095301 UP000268350 UP000192221 UP000001819 UP000092445 UP000092553 UP000078542 UP000005205 UP000092444 UP000007801 UP000076858 UP000007755 UP000078492 UP000091820 UP000078540 UP000053105 UP000215335 UP000002358 UP000000305 UP000014760 UP000001554 UP000261480 UP000261540 UP000225706 UP000261440 UP000008237 UP000000437 UP000221080 UP000016666 UP000275408 UP000028760 UP000078200 UP000087266 UP000007267 UP000261500 UP000053454 UP000002852 UP000008143 UP000018467 UP000265140 UP000189705 UP000053872
UP000002320 UP000019118 UP000001070 UP000030742 UP000095300 UP000076502 UP000008792 UP000000803 UP000000304 UP000002282 UP000279307 UP000008711 UP000001292 UP000007798 UP000037069 UP000092443 UP000009192 UP000009046 UP000092460 UP000008820 UP000095301 UP000268350 UP000192221 UP000001819 UP000092445 UP000092553 UP000078542 UP000005205 UP000092444 UP000007801 UP000076858 UP000007755 UP000078492 UP000091820 UP000078540 UP000053105 UP000215335 UP000002358 UP000000305 UP000014760 UP000001554 UP000261480 UP000261540 UP000225706 UP000261440 UP000008237 UP000000437 UP000221080 UP000016666 UP000275408 UP000028760 UP000078200 UP000087266 UP000007267 UP000261500 UP000053454 UP000002852 UP000008143 UP000018467 UP000265140 UP000189705 UP000053872
ProteinModelPortal
H9IYT9
A0A2A4JEZ5
A0A194QBG4
A0A212EHR8
A0A2H1WJZ4
A0A2A4IT09
+ More
A0A336MJ09 A0A336L450 A0A067RMR1 A0A1L8DME8 A0A0K8TKB2 D6X4C6 A0A1Y1JXA6 B0W7H5 A0A1Q3F798 N6UE47 B4JVA3 U4U056 A0A1I8PTP5 W8BLD5 A0A154PU72 B4LYM2 A0A034VZX0 A0A0A9XYL0 A0A0K8UQY4 A0A0A1X9R0 Q9VA71 B4R1M5 Q8T8R5 B4PLW8 A0A3L8DD54 B3P585 B4HZR2 B4NGW0 A0A0L0BLE6 A0A1A9XTD2 B4K8R5 E0VIA9 A0A1B0BUR2 Q176G0 A0A1I8MC82 A0A3B0K3A1 A0A1W4W458 Q29C42 A0A1A9ZZK4 A0A0M3QYH5 A0A151IQG2 A0A158NU93 A0A1B0FR88 B3MTC0 A0A0P5ECE0 F4WR70 A0A151JCT4 A0A1A9WN38 A0A1B6DP92 A0A195BBN9 A0A0N0BF55 A0A0P4WA17 A0A232FC62 K7J2B7 E9H3M7 R7UEM3 A0A0P5YAE6 A0A1B6G315 C3XZW8 A0A0P6IFD4 A0A3B3YK73 A0A1Z5L619 A0A1B6HV73 A0A293LZQ2 A0A3B3QTS4 A0A2B4RMA9 A0A3B4EER2 E2BU25 Q6TH47 A0A2D0QZI6 F1QBG8 A0A2R5LNF0 A0A3B3X5Z8 U3ID15 A0A3M6V5R6 A0A087XLE7 A0A1A9VVI2 A0A1S3PKR0 R0L8X6 K7FH03 A0A3B3UUW0 A0A0L8GQP8 A0A3N0Z715 A0A0L8GRS4 M4AM50 F7EMR4 W5KDA0 A0A3P8Y729 K7FH21 A0A1A7X4E7 A0A1U7RXZ4 V9KWF6 R7VTM2 A0A093J235
A0A336MJ09 A0A336L450 A0A067RMR1 A0A1L8DME8 A0A0K8TKB2 D6X4C6 A0A1Y1JXA6 B0W7H5 A0A1Q3F798 N6UE47 B4JVA3 U4U056 A0A1I8PTP5 W8BLD5 A0A154PU72 B4LYM2 A0A034VZX0 A0A0A9XYL0 A0A0K8UQY4 A0A0A1X9R0 Q9VA71 B4R1M5 Q8T8R5 B4PLW8 A0A3L8DD54 B3P585 B4HZR2 B4NGW0 A0A0L0BLE6 A0A1A9XTD2 B4K8R5 E0VIA9 A0A1B0BUR2 Q176G0 A0A1I8MC82 A0A3B0K3A1 A0A1W4W458 Q29C42 A0A1A9ZZK4 A0A0M3QYH5 A0A151IQG2 A0A158NU93 A0A1B0FR88 B3MTC0 A0A0P5ECE0 F4WR70 A0A151JCT4 A0A1A9WN38 A0A1B6DP92 A0A195BBN9 A0A0N0BF55 A0A0P4WA17 A0A232FC62 K7J2B7 E9H3M7 R7UEM3 A0A0P5YAE6 A0A1B6G315 C3XZW8 A0A0P6IFD4 A0A3B3YK73 A0A1Z5L619 A0A1B6HV73 A0A293LZQ2 A0A3B3QTS4 A0A2B4RMA9 A0A3B4EER2 E2BU25 Q6TH47 A0A2D0QZI6 F1QBG8 A0A2R5LNF0 A0A3B3X5Z8 U3ID15 A0A3M6V5R6 A0A087XLE7 A0A1A9VVI2 A0A1S3PKR0 R0L8X6 K7FH03 A0A3B3UUW0 A0A0L8GQP8 A0A3N0Z715 A0A0L8GRS4 M4AM50 F7EMR4 W5KDA0 A0A3P8Y729 K7FH21 A0A1A7X4E7 A0A1U7RXZ4 V9KWF6 R7VTM2 A0A093J235
PDB
3RZU
E-value=1.71086e-56,
Score=555
Ontologies
GO
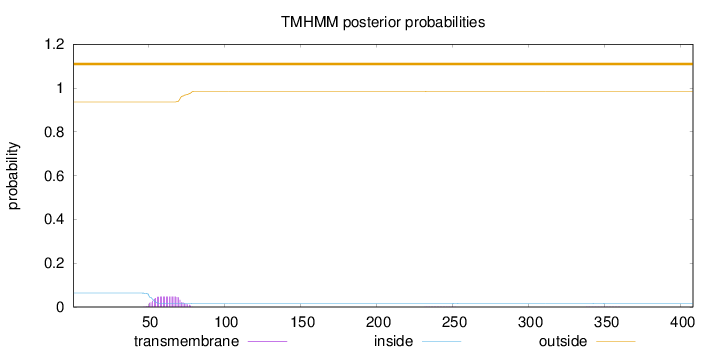
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00728
Exp number, first 60 AAs:
0.38842
Total prob of N-in:
0.06374
outside
1 - 408
Population Genetic Test Statistics
Pi
298.247527
Theta
182.384592
Tajima's D
1.667095
CLR
0.006465
CSRT
0.822958852057397
Interpretation
Uncertain
