Pre Gene Modal
BGIBMGA002486
Annotation
PREDICTED:_mannosyl-oligosaccharide_alpha-1?2-mannosidase_isoform_A_isoform_X2_[Bombyx_mori]
Full name
alpha-1,2-Mannosidase
+ More
Mannosyl-oligosaccharide alpha-1,2-mannosidase IA
Mannosyl-oligosaccharide alpha-1,2-mannosidase IA
Alternative Name
Man(9)-alpha-mannosidase
Mannosidase-1
Mannosidase-1
Location in the cell
Cytoplasmic Reliability : 1.095 Mitochondrial Reliability : 1.058
Sequence
CDS
ATGCGTGCTTATCCCCGTTTGATTTTGACCGCGGTCACTGCTATTATCTGTATGAGTTTGCTTGTCCAAACATTCAACGCGGGACAGGGAACATCGGTGCATCCAGCTAGTCACCTGGCCCGCGAGGCCCGATCACTTCAATCTAACCAAATCGAAGCTGGTAACAGCAGCAGCTACAGCGGAGCGTTAGATTCCCAGTCATCAGTCGAAAAACAGCGGTACATTATCGAGATGATGCGACACGCTTGGAACAATTACAAACTGTACGCTTGGGGTAAAAATGAACTGAAACCGATATCTAAACGCGCACATTTGTCGAGCGTGTTTGGCTCCGGGGAGATGGGAGCCACCATTGTTGACGGCCTGGACACCTTGTACGTGATGGGCCTGACCGACGAGTTCCGTGAGGGACGTGACTGGATCGCGGAACATTTCAACATCAACGAAATCGATTCCGAACTATCAGTATTCGAAACGACGATACGTTTTGTGGGAGGTCTGCTGTCTTGCTACGCTCTGACCGGTGACACCATCTTCAGAGACAAAGCCGCCGAGGTCGCCGACGCCCTGCTGCCTGCCTTCGAAACTCCCACTGGACTCCCATACGCTTTAATCAATCCATCCACTAAGGCCAGCCGCCAGTATCACTGGGCAGGCCCGAACAGTATCCTCTCTGAATTGGGTACATTGCACCTCGAGTTCTCATACCTGAGTGACGTCACAGGACGTGAAGTCTACAGGAACAAAGTGGATAAAATACGTGAAGTTCTCTACAACATTGAGAAGCCCGGTGATCTTTATCCGAACTTCATCAATCCTCGAACCGGACAGTGGGGACAAAGACACATATCCCTGGGAGCTCTGGGCGACTCGTTCTACGAGTACCTGCTGAAGGCCTGGCTCGCGTCGAACCGCGGCGACGAGCAGGCGCGCACCATGTTCGACACGGCGATGCAGGCCGCGCTCGACAAGATGCTGCGCGTGTCGCCCTCCGGCCTCGCCTACCTCGCCGAACTCAAGTACGGCAGGGTCATCGAAGAGAAGATGGATCACTTGTCTTGCTTCGCTGGTGGCATGTTCGCTTTGGCCGCTACGACATTGGAGAATTCGTTGTCTGAACGGTACATGGACGTCGCAAGGAAACTGACCCACACCTGCCACGAGTCCTACTTCAGATCCGAAACTAAACTGGGACCAGAGGCATTCAGATTTTCGAACGCAGTCGAAGCCCGAGCTCAGAAGAGCAACGAGAAAGTTTACATACTGAGGCCGGAGACGTTCGAGAGCTACTTCATCATGTGGCGACTCACCAAGGAGCAGAAGTACCGCGACTGGGGCTGGGAGGCGGTTCAGGCGCTAGAGAAGCATTGTCGCGTCGAAGGCGGCTACACCGGACTCACGAACGTCTACCACCAGAACCCTCAGGGCGACGACGTGCAACAGAGCTACTTCCTCGCTGAAACACTTAAGTACCTCTACCTTCTGTTCTCGGACGACTCTCTCCTTTCGCTCGACGAGTGGGTGTTCAACACTGAAGCTCATCCGCTTCCAATTAAAGGCAAGAATCCCCTGTACCGCATGGCGAGCACGCCCGTGCCAATAGAAGACAACCGAGTCTAA
Protein
MRAYPRLILTAVTAIICMSLLVQTFNAGQGTSVHPASHLAREARSLQSNQIEAGNSSSYSGALDSQSSVEKQRYIIEMMRHAWNNYKLYAWGKNELKPISKRAHLSSVFGSGEMGATIVDGLDTLYVMGLTDEFREGRDWIAEHFNINEIDSELSVFETTIRFVGGLLSCYALTGDTIFRDKAAEVADALLPAFETPTGLPYALINPSTKASRQYHWAGPNSILSELGTLHLEFSYLSDVTGREVYRNKVDKIREVLYNIEKPGDLYPNFINPRTGQWGQRHISLGALGDSFYEYLLKAWLASNRGDEQARTMFDTAMQAALDKMLRVSPSGLAYLAELKYGRVIEEKMDHLSCFAGGMFALAATTLENSLSERYMDVARKLTHTCHESYFRSETKLGPEAFRFSNAVEARAQKSNEKVYILRPETFESYFIMWRLTKEQKYRDWGWEAVQALEKHCRVEGGYTGLTNVYHQNPQGDDVQQSYFLAETLKYLYLLFSDDSLLSLDEWVFNTEAHPLPIKGKNPLYRMASTPVPIEDNRV
Summary
Description
Involved in the maturation of Asn-linked oligosaccharides. Progressively trim alpha-1,2-linked mannose residues from Man(9)GlcNAc(2) to produce Man(5)GlcNAc(2).
Catalytic Activity
Hydrolysis of the terminal (1->2)-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2).
Similarity
Belongs to the glycosyl hydrolase 47 family.
Keywords
Alternative promoter usage
Calcium
Complete proteome
Disulfide bond
Glycoprotein
Glycosidase
Golgi apparatus
Hydrolase
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain alpha-1,2-Mannosidase
splice variant In isoform K.
splice variant In isoform K.
Uniprot
H9IZ01
H9IYU1
A0A2H1WJZ7
A0A194QD26
A0A194RNB4
O18498
+ More
A0A212EHT2 B4JXQ3 P53624 A0A139WAC4 A0A139WAB1 P53624-2 A0A1Y1KXM7 B4IDP0 A0A0Q5TDT9 B3NW46 Q8IRL4 A0A1I8MGL1 A0A0Q9WMV7 B4PXH4 A0A0Q5THZ9 A0A0R1ECD8 A0A1Y1KZY0 A0A0R1EBX4 T1P828 B4L336 A0A0Q9WPD3 A0A0Q9WM44 A0A2J7QK67 B3MW69 A0A1W4VUH6 A0A1I8PYM2 A0A1W4VTQ6 V5H014 A0A1B0C9F4 A0A1W4VG52 A0A3B0JZ12 B4NDR0 B4M3C5 A0A0M4EUG7 A0A084VDD7 A0A1I8MQ91 A0A1I8PYK5 A0A182IR84 A0A0L0C1T1 U4UXU6 Q29CI1 A0A1J1HHC9 A0A1J1HHC8 A0A1L8DR25 E0VI77 A0A067R784 U5EW55 A0A1B0FJA7 A0A0V0G9J1 A0A0K8UXX6 B4H0T3 A0A0K8UCW3 A0A336LLS7 A0A0A1WR15 A0A069DWB1 A0A182FK62 A0A224XK05 A0A0P4W0H0 A0A1A9XXP1 A0A1A9VY99 A0A1B0APE7 A0A1Q3FXV6 A0A1Q3FXU1 A0A0T6B9R8 A0A1B0ADR1 A0A1Q3G2C4 A0A1Q3G2D8 A0A146M1M6 A0A0A9X5C5 A0A1W4WPU2 A0A1A9W8L9 A0A0P6J5H2 A0A2M4BIY6 A0A2M4BFT9 A0A2M4CSW7 A0A2M3YYF6 A0A2M4AMB6 A0A2M4A8U7 A0A2M3Z0S8 A0A2M4A6C9 A0A2M4ACR5 A0A2M4A694 A0A2M4A6G7 B0WSW5 A0A1S4G324 A0A2M4CS71 A0A2M4CS39 Q16FK8 A0A182N1H4 E9FX86 A0A182GD08 A0A1Y1L4V4 A0A182VD90 Q16IH0
A0A212EHT2 B4JXQ3 P53624 A0A139WAC4 A0A139WAB1 P53624-2 A0A1Y1KXM7 B4IDP0 A0A0Q5TDT9 B3NW46 Q8IRL4 A0A1I8MGL1 A0A0Q9WMV7 B4PXH4 A0A0Q5THZ9 A0A0R1ECD8 A0A1Y1KZY0 A0A0R1EBX4 T1P828 B4L336 A0A0Q9WPD3 A0A0Q9WM44 A0A2J7QK67 B3MW69 A0A1W4VUH6 A0A1I8PYM2 A0A1W4VTQ6 V5H014 A0A1B0C9F4 A0A1W4VG52 A0A3B0JZ12 B4NDR0 B4M3C5 A0A0M4EUG7 A0A084VDD7 A0A1I8MQ91 A0A1I8PYK5 A0A182IR84 A0A0L0C1T1 U4UXU6 Q29CI1 A0A1J1HHC9 A0A1J1HHC8 A0A1L8DR25 E0VI77 A0A067R784 U5EW55 A0A1B0FJA7 A0A0V0G9J1 A0A0K8UXX6 B4H0T3 A0A0K8UCW3 A0A336LLS7 A0A0A1WR15 A0A069DWB1 A0A182FK62 A0A224XK05 A0A0P4W0H0 A0A1A9XXP1 A0A1A9VY99 A0A1B0APE7 A0A1Q3FXV6 A0A1Q3FXU1 A0A0T6B9R8 A0A1B0ADR1 A0A1Q3G2C4 A0A1Q3G2D8 A0A146M1M6 A0A0A9X5C5 A0A1W4WPU2 A0A1A9W8L9 A0A0P6J5H2 A0A2M4BIY6 A0A2M4BFT9 A0A2M4CSW7 A0A2M3YYF6 A0A2M4AMB6 A0A2M4A8U7 A0A2M3Z0S8 A0A2M4A6C9 A0A2M4ACR5 A0A2M4A694 A0A2M4A6G7 B0WSW5 A0A1S4G324 A0A2M4CS71 A0A2M4CS39 Q16FK8 A0A182N1H4 E9FX86 A0A182GD08 A0A1Y1L4V4 A0A182VD90 Q16IH0
EC Number
3.2.1.-
3.2.1.113
3.2.1.113
Pubmed
19121390
26354079
9147053
22118469
17994087
7729592
+ More
10731132 12537572 18362917 19820115 28004739 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 18057021 24438588 26108605 23537049 15632085 20566863 24845553 25830018 26334808 27129103 26823975 25401762 26999592 17510324 21292972 26483478
10731132 12537572 18362917 19820115 28004739 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 18057021 24438588 26108605 23537049 15632085 20566863 24845553 25830018 26334808 27129103 26823975 25401762 26999592 17510324 21292972 26483478
EMBL
BABH01010832
BABH01010836
BABH01010837
ODYU01009163
SOQ53401.1
KQ459220
+ More
KPJ02880.1 KQ459984 KPJ19002.1 AF005035 AAB62720.1 AGBW02014807 OWR41031.1 CH916376 EDV95152.1 X82640 X82641 AE014298 BT011036 KQ971410 KYB24856.1 KYB24855.1 GEZM01071031 JAV66112.1 CH480830 EDW45698.1 CH954180 KQS29861.1 EDV46179.1 BT024451 AAN09256.2 ABC86513.1 CH933810 KRF93895.1 CM000162 EDX02925.1 KQS29860.1 KRK07021.1 GEZM01071035 JAV66108.1 KRK07020.1 KA644704 AFP59333.1 EDW06964.1 CH940651 KRF82176.1 KRF82175.1 NEVH01013277 PNF28973.1 CH902625 EDV35214.1 GALX01002318 JAB66148.1 AJWK01002278 OUUW01000011 SPP86313.1 CH964239 EDW81879.1 EDW65300.1 KRF82174.1 CP012528 ALC49401.1 ATLV01011268 ATLV01011269 KE524660 KFB35981.1 JRES01000996 KNC26313.1 KB632410 ERL95186.1 CH475471 EAL29254.3 CVRI01000004 CRK87431.1 CRK87432.1 GFDF01005260 JAV08824.1 DS235184 EEB13083.1 KK852898 KDR14159.1 GANO01001574 JAB58297.1 CCAG010011096 CCAG010011097 GECL01001616 JAP04508.1 GDHF01020931 JAI31383.1 CH479201 EDW29998.1 GDHF01028114 GDHF01008468 JAI24200.1 JAI43846.1 UFQT01000005 SSX17317.1 GBXI01013332 JAD00960.1 GBGD01000758 JAC88131.1 GFTR01007494 JAW08932.1 GDKW01000233 JAI56362.1 JXJN01001384 GFDL01002638 JAV32407.1 GFDL01002648 JAV32397.1 LJIG01002912 KRT84028.1 GFDL01001067 JAV33978.1 GFDL01001060 JAV33985.1 GDHC01005952 JAQ12677.1 GBHO01031304 GBHO01012830 GBRD01008954 GDHC01013097 JAG12300.1 JAG30774.1 JAG56867.1 JAQ05532.1 GDUN01000555 JAN95364.1 GGFJ01003637 MBW52778.1 GGFJ01002779 MBW51920.1 GGFL01003750 MBW67928.1 GGFM01000534 MBW21285.1 GGFK01008551 MBW41872.1 GGFK01003829 MBW37150.1 GGFM01001386 MBW22137.1 GGFK01002990 MBW36311.1 GGFK01005250 MBW38571.1 GGFK01003003 MBW36324.1 GGFK01003024 MBW36345.1 DS232077 EDS34093.1 GGFL01003989 MBW68167.1 GGFL01003988 MBW68166.1 CH478417 EAT33026.1 GL732526 EFX88311.1 JXUM01055002 JXUM01055003 KQ561847 KXJ77349.1 GEZM01071036 JAV66107.1 CH478078 EAT34060.1
KPJ02880.1 KQ459984 KPJ19002.1 AF005035 AAB62720.1 AGBW02014807 OWR41031.1 CH916376 EDV95152.1 X82640 X82641 AE014298 BT011036 KQ971410 KYB24856.1 KYB24855.1 GEZM01071031 JAV66112.1 CH480830 EDW45698.1 CH954180 KQS29861.1 EDV46179.1 BT024451 AAN09256.2 ABC86513.1 CH933810 KRF93895.1 CM000162 EDX02925.1 KQS29860.1 KRK07021.1 GEZM01071035 JAV66108.1 KRK07020.1 KA644704 AFP59333.1 EDW06964.1 CH940651 KRF82176.1 KRF82175.1 NEVH01013277 PNF28973.1 CH902625 EDV35214.1 GALX01002318 JAB66148.1 AJWK01002278 OUUW01000011 SPP86313.1 CH964239 EDW81879.1 EDW65300.1 KRF82174.1 CP012528 ALC49401.1 ATLV01011268 ATLV01011269 KE524660 KFB35981.1 JRES01000996 KNC26313.1 KB632410 ERL95186.1 CH475471 EAL29254.3 CVRI01000004 CRK87431.1 CRK87432.1 GFDF01005260 JAV08824.1 DS235184 EEB13083.1 KK852898 KDR14159.1 GANO01001574 JAB58297.1 CCAG010011096 CCAG010011097 GECL01001616 JAP04508.1 GDHF01020931 JAI31383.1 CH479201 EDW29998.1 GDHF01028114 GDHF01008468 JAI24200.1 JAI43846.1 UFQT01000005 SSX17317.1 GBXI01013332 JAD00960.1 GBGD01000758 JAC88131.1 GFTR01007494 JAW08932.1 GDKW01000233 JAI56362.1 JXJN01001384 GFDL01002638 JAV32407.1 GFDL01002648 JAV32397.1 LJIG01002912 KRT84028.1 GFDL01001067 JAV33978.1 GFDL01001060 JAV33985.1 GDHC01005952 JAQ12677.1 GBHO01031304 GBHO01012830 GBRD01008954 GDHC01013097 JAG12300.1 JAG30774.1 JAG56867.1 JAQ05532.1 GDUN01000555 JAN95364.1 GGFJ01003637 MBW52778.1 GGFJ01002779 MBW51920.1 GGFL01003750 MBW67928.1 GGFM01000534 MBW21285.1 GGFK01008551 MBW41872.1 GGFK01003829 MBW37150.1 GGFM01001386 MBW22137.1 GGFK01002990 MBW36311.1 GGFK01005250 MBW38571.1 GGFK01003003 MBW36324.1 GGFK01003024 MBW36345.1 DS232077 EDS34093.1 GGFL01003989 MBW68167.1 GGFL01003988 MBW68166.1 CH478417 EAT33026.1 GL732526 EFX88311.1 JXUM01055002 JXUM01055003 KQ561847 KXJ77349.1 GEZM01071036 JAV66107.1 CH478078 EAT34060.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000001070
UP000000803
+ More
UP000007266 UP000001292 UP000008711 UP000095301 UP000009192 UP000002282 UP000008792 UP000235965 UP000007801 UP000192221 UP000095300 UP000092461 UP000268350 UP000007798 UP000092553 UP000030765 UP000075880 UP000037069 UP000030742 UP000001819 UP000183832 UP000009046 UP000027135 UP000092444 UP000008744 UP000069272 UP000092443 UP000078200 UP000092460 UP000092445 UP000192223 UP000091820 UP000002320 UP000008820 UP000075884 UP000000305 UP000069940 UP000249989 UP000075903
UP000007266 UP000001292 UP000008711 UP000095301 UP000009192 UP000002282 UP000008792 UP000235965 UP000007801 UP000192221 UP000095300 UP000092461 UP000268350 UP000007798 UP000092553 UP000030765 UP000075880 UP000037069 UP000030742 UP000001819 UP000183832 UP000009046 UP000027135 UP000092444 UP000008744 UP000069272 UP000092443 UP000078200 UP000092460 UP000092445 UP000192223 UP000091820 UP000002320 UP000008820 UP000075884 UP000000305 UP000069940 UP000249989 UP000075903
Pfam
PF01532 Glyco_hydro_47
Interpro
SUPFAM
SSF48225
SSF48225
Gene 3D
ProteinModelPortal
H9IZ01
H9IYU1
A0A2H1WJZ7
A0A194QD26
A0A194RNB4
O18498
+ More
A0A212EHT2 B4JXQ3 P53624 A0A139WAC4 A0A139WAB1 P53624-2 A0A1Y1KXM7 B4IDP0 A0A0Q5TDT9 B3NW46 Q8IRL4 A0A1I8MGL1 A0A0Q9WMV7 B4PXH4 A0A0Q5THZ9 A0A0R1ECD8 A0A1Y1KZY0 A0A0R1EBX4 T1P828 B4L336 A0A0Q9WPD3 A0A0Q9WM44 A0A2J7QK67 B3MW69 A0A1W4VUH6 A0A1I8PYM2 A0A1W4VTQ6 V5H014 A0A1B0C9F4 A0A1W4VG52 A0A3B0JZ12 B4NDR0 B4M3C5 A0A0M4EUG7 A0A084VDD7 A0A1I8MQ91 A0A1I8PYK5 A0A182IR84 A0A0L0C1T1 U4UXU6 Q29CI1 A0A1J1HHC9 A0A1J1HHC8 A0A1L8DR25 E0VI77 A0A067R784 U5EW55 A0A1B0FJA7 A0A0V0G9J1 A0A0K8UXX6 B4H0T3 A0A0K8UCW3 A0A336LLS7 A0A0A1WR15 A0A069DWB1 A0A182FK62 A0A224XK05 A0A0P4W0H0 A0A1A9XXP1 A0A1A9VY99 A0A1B0APE7 A0A1Q3FXV6 A0A1Q3FXU1 A0A0T6B9R8 A0A1B0ADR1 A0A1Q3G2C4 A0A1Q3G2D8 A0A146M1M6 A0A0A9X5C5 A0A1W4WPU2 A0A1A9W8L9 A0A0P6J5H2 A0A2M4BIY6 A0A2M4BFT9 A0A2M4CSW7 A0A2M3YYF6 A0A2M4AMB6 A0A2M4A8U7 A0A2M3Z0S8 A0A2M4A6C9 A0A2M4ACR5 A0A2M4A694 A0A2M4A6G7 B0WSW5 A0A1S4G324 A0A2M4CS71 A0A2M4CS39 Q16FK8 A0A182N1H4 E9FX86 A0A182GD08 A0A1Y1L4V4 A0A182VD90 Q16IH0
A0A212EHT2 B4JXQ3 P53624 A0A139WAC4 A0A139WAB1 P53624-2 A0A1Y1KXM7 B4IDP0 A0A0Q5TDT9 B3NW46 Q8IRL4 A0A1I8MGL1 A0A0Q9WMV7 B4PXH4 A0A0Q5THZ9 A0A0R1ECD8 A0A1Y1KZY0 A0A0R1EBX4 T1P828 B4L336 A0A0Q9WPD3 A0A0Q9WM44 A0A2J7QK67 B3MW69 A0A1W4VUH6 A0A1I8PYM2 A0A1W4VTQ6 V5H014 A0A1B0C9F4 A0A1W4VG52 A0A3B0JZ12 B4NDR0 B4M3C5 A0A0M4EUG7 A0A084VDD7 A0A1I8MQ91 A0A1I8PYK5 A0A182IR84 A0A0L0C1T1 U4UXU6 Q29CI1 A0A1J1HHC9 A0A1J1HHC8 A0A1L8DR25 E0VI77 A0A067R784 U5EW55 A0A1B0FJA7 A0A0V0G9J1 A0A0K8UXX6 B4H0T3 A0A0K8UCW3 A0A336LLS7 A0A0A1WR15 A0A069DWB1 A0A182FK62 A0A224XK05 A0A0P4W0H0 A0A1A9XXP1 A0A1A9VY99 A0A1B0APE7 A0A1Q3FXV6 A0A1Q3FXU1 A0A0T6B9R8 A0A1B0ADR1 A0A1Q3G2C4 A0A1Q3G2D8 A0A146M1M6 A0A0A9X5C5 A0A1W4WPU2 A0A1A9W8L9 A0A0P6J5H2 A0A2M4BIY6 A0A2M4BFT9 A0A2M4CSW7 A0A2M3YYF6 A0A2M4AMB6 A0A2M4A8U7 A0A2M3Z0S8 A0A2M4A6C9 A0A2M4ACR5 A0A2M4A694 A0A2M4A6G7 B0WSW5 A0A1S4G324 A0A2M4CS71 A0A2M4CS39 Q16FK8 A0A182N1H4 E9FX86 A0A182GD08 A0A1Y1L4V4 A0A182VD90 Q16IH0
PDB
5KKB
E-value=2.02919e-144,
Score=1315
Ontologies
GO
Topology
Subcellular location
Golgi apparatus membrane
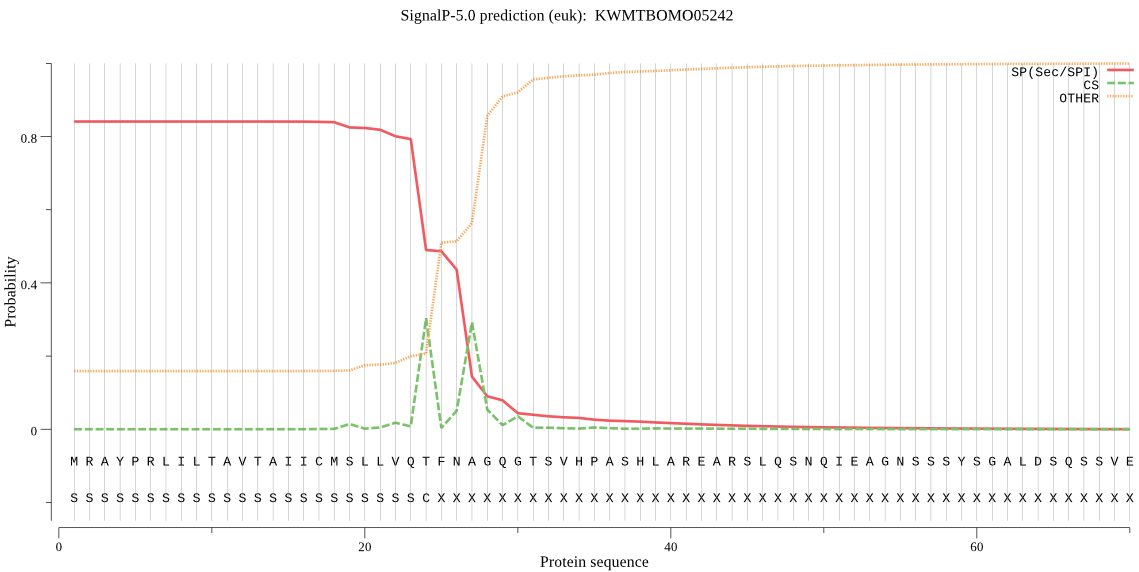
SignalP
Position: 1 - 24,
Likelihood: 0.840795
Length:
539
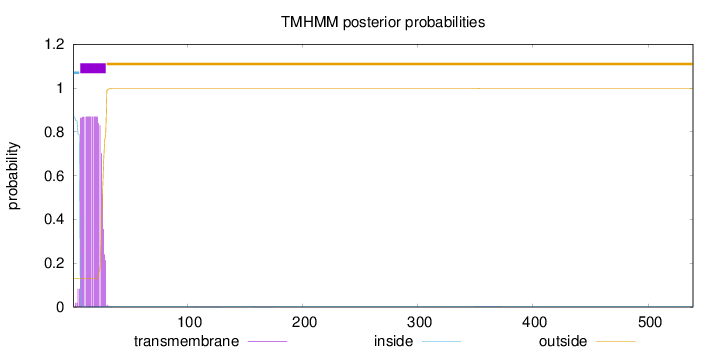
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.8818799999999
Exp number, first 60 AAs:
17.82193
Total prob of N-in:
0.86994
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 539
Population Genetic Test Statistics
Pi
222.152756
Theta
179.338326
Tajima's D
0.996069
CLR
0.002785
CSRT
0.658767061646918
Interpretation
Uncertain
