Gene
KWMTBOMO05240 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002485
Annotation
alpha_1?2-mannosidase_[Spodoptera_frugiperda]
Full name
alpha-1,2-Mannosidase
Location in the cell
Mitochondrial Reliability : 1
Sequence
CDS
ATGACGGGAATTTTACCTACGTATCAGCGGTTTGTTAACGGGGTACCCGTCCCGTCCATATCTCGGCGGTCGTTCCGTATAAGAGAGAAATATCTGATTGTTTCTGTACTTTTGACGTTCGGAATCGTGTGGTTAGGAGCTCTGTTTTATTTACCTGAATTTAAAAGTTCGAGCAGTGTGAATGACAGTGTTTACAATGTGTATAAACGTATTCAGAAAGCGGGTCCAGAGCTGTTAATGCCTCCGCCACTCGCTCAAAACGATGTGGGAGACTTTCCTGTAGTCGGTATCGTTCGGCATGGCGAAGAAGGTGACGACCCACACATAGTTGAGGACAGAAACAGACTTAAGGCTAAGATAGACGAAGACATGGGCATGAAAGTTCTCGAGAGACCGCAGTTTGATGTTGCACCTTCTGTATCTTCTTCTCGAGGTCCCAGCAAGCCTCCTGTTGACGCAATAGAGGAGCCCGCTGTTAGGAACTTCGCAGCTAAAGACGCATCTCCTGCTGGTCCTAAACCAGACGGGTCTAATAGATACGTCGCTGTTGCTATCGGTCCTAACGCTGATCCAGATCAGAAACAAAAGTTAGAGACAGTTAAAGAGTAA
Protein
MTGILPTYQRFVNGVPVPSISRRSFRIREKYLIVSVLLTFGIVWLGALFYLPEFKSSSSVNDSVYNVYKRIQKAGPELLMPPPLAQNDVGDFPVVGIVRHGEEGDDPHIVEDRNRLKAKIDEDMGMKVLERPQFDVAPSVSSSRGPSKPPVDAIEEPAVRNFAAKDASPAGPKPDGSNRYVAVAIGPNADPDQKQKLETVKE
Summary
Similarity
Belongs to the glycosyl hydrolase 47 family.
Uniprot
H9IZ00
A0A2H1WJZ0
O18498
A0A194QBT4
A0A194RSF6
A0A212FB20
+ More
A0A0L7KZQ7 A0A2P8YN80 A0A1Y1L020 A0A1Y1KXH0 A0A1Y1KZY0 A0A1B6EL18 A0A0T6B092 A0A1B6MAL9 A0A1B6H5E4 A0A2J7QK79 A0A1B6D8W2 A0A0A9YCH2 A0A0P4W0H0 A0A069DWB1 A0A0A9X5C5 E2BAD4 A0A139WAB1 A0A139WAC4 V5H014 A0A2R7WBG6 A0A195BLC2 A0A087ZY30 A0A0L7R8Y0 A0A2A3EPS3 A0A026VVZ3 A0A3L8DBC9 A0A154PF35 A0A0C9Q8X5 A0A067R784 A0A232F7V8 A0A023GL76 A0A1E1X7K4 A0A131YJA8 L7M7E3 A0A131XF80 T1IIC1 A0A2R5LN53 A0A1W7RB10 A0A2M4BFT9 A0A1B0C9F4 A0A0P6A5S6 E9FX86 A0A0P4VWX6 A0A0P4W3H4 A0A0P6HHC7 A0A0P5PD52 A0A0P5ZD43 A0A0P5HGD1 A0A0P5C4G3 A0A0P5CEV0 A0A0P5HQP0 A0A0P5CH63 A0A0N8B595
A0A0L7KZQ7 A0A2P8YN80 A0A1Y1L020 A0A1Y1KXH0 A0A1Y1KZY0 A0A1B6EL18 A0A0T6B092 A0A1B6MAL9 A0A1B6H5E4 A0A2J7QK79 A0A1B6D8W2 A0A0A9YCH2 A0A0P4W0H0 A0A069DWB1 A0A0A9X5C5 E2BAD4 A0A139WAB1 A0A139WAC4 V5H014 A0A2R7WBG6 A0A195BLC2 A0A087ZY30 A0A0L7R8Y0 A0A2A3EPS3 A0A026VVZ3 A0A3L8DBC9 A0A154PF35 A0A0C9Q8X5 A0A067R784 A0A232F7V8 A0A023GL76 A0A1E1X7K4 A0A131YJA8 L7M7E3 A0A131XF80 T1IIC1 A0A2R5LN53 A0A1W7RB10 A0A2M4BFT9 A0A1B0C9F4 A0A0P6A5S6 E9FX86 A0A0P4VWX6 A0A0P4W3H4 A0A0P6HHC7 A0A0P5PD52 A0A0P5ZD43 A0A0P5HGD1 A0A0P5C4G3 A0A0P5CEV0 A0A0P5HQP0 A0A0P5CH63 A0A0N8B595
EC Number
3.2.1.-
Pubmed
EMBL
BABH01010823
ODYU01009163
SOQ53400.1
AF005035
AAB62720.1
KQ459220
+ More
KPJ02879.1 KQ459984 KPJ19001.1 AGBW02009383 OWR50942.1 JTDY01003990 KOB68743.1 PYGN01000474 PSN45696.1 GEZM01071032 JAV66111.1 GEZM01071034 JAV66109.1 GEZM01071035 JAV66108.1 GECZ01031154 JAS38615.1 LJIG01016382 KRT80769.1 GEBQ01007073 JAT32904.1 GECU01037813 JAS69893.1 NEVH01013277 PNF28987.1 GEDC01015171 JAS22127.1 GBHO01012827 JAG30777.1 GDKW01000233 JAI56362.1 GBGD01000758 JAC88131.1 GBHO01031304 GBHO01012830 GBRD01008954 GDHC01013097 JAG12300.1 JAG30774.1 JAG56867.1 JAQ05532.1 GL446702 EFN87348.1 KQ971410 KYB24855.1 KYB24856.1 GALX01002318 JAB66148.1 KK854495 PTY16350.1 KQ976453 KYM85477.1 KQ414629 KOC67299.1 KZ288197 PBC33755.1 KK107746 EZA47932.1 QOIP01000010 RLU17413.1 KQ434878 KZC09918.1 GBYB01010728 JAG80495.1 KK852898 KDR14159.1 NNAY01000783 OXU26533.1 GBBM01000839 JAC34579.1 GFAC01003939 JAT95249.1 GEDV01010401 JAP78156.1 GACK01005064 JAA59970.1 GEFH01003569 JAP65012.1 JH430159 GGLE01006837 MBY10963.1 GFAH01000059 JAV48330.1 GGFJ01002779 MBW51920.1 AJWK01002278 GDIP01033896 JAM69819.1 GL732526 EFX88311.1 GDRN01090731 JAI60423.1 GDRN01090732 JAI60422.1 GDIQ01018849 JAN75888.1 GDIQ01134368 JAL17358.1 GDIP01045708 JAM58007.1 GDIQ01230085 JAK21640.1 GDIP01191383 JAJ32019.1 GDIP01171595 JAJ51807.1 GDIQ01230086 JAK21639.1 GDIP01171596 JAJ51806.1 GDIQ01211499 JAK40226.1
KPJ02879.1 KQ459984 KPJ19001.1 AGBW02009383 OWR50942.1 JTDY01003990 KOB68743.1 PYGN01000474 PSN45696.1 GEZM01071032 JAV66111.1 GEZM01071034 JAV66109.1 GEZM01071035 JAV66108.1 GECZ01031154 JAS38615.1 LJIG01016382 KRT80769.1 GEBQ01007073 JAT32904.1 GECU01037813 JAS69893.1 NEVH01013277 PNF28987.1 GEDC01015171 JAS22127.1 GBHO01012827 JAG30777.1 GDKW01000233 JAI56362.1 GBGD01000758 JAC88131.1 GBHO01031304 GBHO01012830 GBRD01008954 GDHC01013097 JAG12300.1 JAG30774.1 JAG56867.1 JAQ05532.1 GL446702 EFN87348.1 KQ971410 KYB24855.1 KYB24856.1 GALX01002318 JAB66148.1 KK854495 PTY16350.1 KQ976453 KYM85477.1 KQ414629 KOC67299.1 KZ288197 PBC33755.1 KK107746 EZA47932.1 QOIP01000010 RLU17413.1 KQ434878 KZC09918.1 GBYB01010728 JAG80495.1 KK852898 KDR14159.1 NNAY01000783 OXU26533.1 GBBM01000839 JAC34579.1 GFAC01003939 JAT95249.1 GEDV01010401 JAP78156.1 GACK01005064 JAA59970.1 GEFH01003569 JAP65012.1 JH430159 GGLE01006837 MBY10963.1 GFAH01000059 JAV48330.1 GGFJ01002779 MBW51920.1 AJWK01002278 GDIP01033896 JAM69819.1 GL732526 EFX88311.1 GDRN01090731 JAI60423.1 GDRN01090732 JAI60422.1 GDIQ01018849 JAN75888.1 GDIQ01134368 JAL17358.1 GDIP01045708 JAM58007.1 GDIQ01230085 JAK21640.1 GDIP01191383 JAJ32019.1 GDIP01171595 JAJ51807.1 GDIQ01230086 JAK21639.1 GDIP01171596 JAJ51806.1 GDIQ01211499 JAK40226.1
Proteomes
PRIDE
Pfam
PF01532 Glyco_hydro_47
Interpro
SUPFAM
SSF48225
SSF48225
Gene 3D
ProteinModelPortal
H9IZ00
A0A2H1WJZ0
O18498
A0A194QBT4
A0A194RSF6
A0A212FB20
+ More
A0A0L7KZQ7 A0A2P8YN80 A0A1Y1L020 A0A1Y1KXH0 A0A1Y1KZY0 A0A1B6EL18 A0A0T6B092 A0A1B6MAL9 A0A1B6H5E4 A0A2J7QK79 A0A1B6D8W2 A0A0A9YCH2 A0A0P4W0H0 A0A069DWB1 A0A0A9X5C5 E2BAD4 A0A139WAB1 A0A139WAC4 V5H014 A0A2R7WBG6 A0A195BLC2 A0A087ZY30 A0A0L7R8Y0 A0A2A3EPS3 A0A026VVZ3 A0A3L8DBC9 A0A154PF35 A0A0C9Q8X5 A0A067R784 A0A232F7V8 A0A023GL76 A0A1E1X7K4 A0A131YJA8 L7M7E3 A0A131XF80 T1IIC1 A0A2R5LN53 A0A1W7RB10 A0A2M4BFT9 A0A1B0C9F4 A0A0P6A5S6 E9FX86 A0A0P4VWX6 A0A0P4W3H4 A0A0P6HHC7 A0A0P5PD52 A0A0P5ZD43 A0A0P5HGD1 A0A0P5C4G3 A0A0P5CEV0 A0A0P5HQP0 A0A0P5CH63 A0A0N8B595
A0A0L7KZQ7 A0A2P8YN80 A0A1Y1L020 A0A1Y1KXH0 A0A1Y1KZY0 A0A1B6EL18 A0A0T6B092 A0A1B6MAL9 A0A1B6H5E4 A0A2J7QK79 A0A1B6D8W2 A0A0A9YCH2 A0A0P4W0H0 A0A069DWB1 A0A0A9X5C5 E2BAD4 A0A139WAB1 A0A139WAC4 V5H014 A0A2R7WBG6 A0A195BLC2 A0A087ZY30 A0A0L7R8Y0 A0A2A3EPS3 A0A026VVZ3 A0A3L8DBC9 A0A154PF35 A0A0C9Q8X5 A0A067R784 A0A232F7V8 A0A023GL76 A0A1E1X7K4 A0A131YJA8 L7M7E3 A0A131XF80 T1IIC1 A0A2R5LN53 A0A1W7RB10 A0A2M4BFT9 A0A1B0C9F4 A0A0P6A5S6 E9FX86 A0A0P4VWX6 A0A0P4W3H4 A0A0P6HHC7 A0A0P5PD52 A0A0P5ZD43 A0A0P5HGD1 A0A0P5C4G3 A0A0P5CEV0 A0A0P5HQP0 A0A0P5CH63 A0A0N8B595
Ontologies
PATHWAY
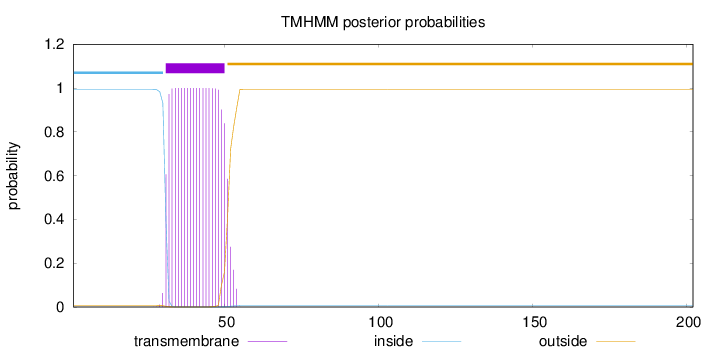
Topology
Length:
202
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.49228
Exp number, first 60 AAs:
20.49209
Total prob of N-in:
0.99365
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 50
outside
51 - 202
Population Genetic Test Statistics
Pi
66.224557
Theta
90.807956
Tajima's D
-0.869354
CLR
704.871308
CSRT
0.16119194040298
Interpretation
Uncertain
