Gene
KWMTBOMO05238
Annotation
PREDICTED:_uncharacterized_protein_LOC106717337_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.839
Sequence
CDS
ATGACCAAAATTTTAATTTTGAAACAAAATGAAAAAATTGCTCTAGTAAAATACGCTAATATATATGATTTGCTAGTGGAGCCATGTGGCATTTTCATTAGCCAAGAGCGCCCATATCTTGCTGCATCTCCTGATGGTGTGTTGGGCGAAGAAACTATAATTGAAGTGAAATGTCCATACGCTAACAGAAAACATGAAATAAATATGCCTTACTTAAAACAATGTAACGGGATATTGTCTCTAAAAAAACCTGACCATATTATTATCAAATCCAAGGGCAGTTATACTGCTCAGGCAAAACATACTGCAACTTGGTCATTTACACCTACAAGGACATCAAAGTGA
Protein
MTKILILKQNEKIALVKYANIYDLLVEPCGIFISQERPYLAASPDGVLGEETIIEVKCPYANRKHEINMPYLKQCNGILSLKKPDHIIIKSKGSYTAQAKHTATWSFTPTRTSK
Summary
Uniprot
A0A1W5BJ07
A0A2S2PNI2
A0A2S2NXS1
A0A1E1XL94
J9M9C6
A0A2S2QHC7
+ More
X1X3F6 J9KTJ5 J9M882 A0A1S3KDQ1 A0A151IKZ0 A0A0J7KCJ1 A0A2H9T450 A0A151JP94 X1XR94 A0A151I918 A0A2S2PJX4 J9L496 V9KQC4 A0A2S2P913 A0A2S2NDB3 A0A164G841 A0A151WZM5 A0A2S2PE96 A0A2S2QV10 A0A026X111 X1WN45 J9M6P6 A0A026W0I6 K7JK45 A0A151J7I0 A0A1X7VJ44 V5G7Y9 A0A1Y1M7F9 A0A2S2RBA8 A0A1Y1LRB1
X1X3F6 J9KTJ5 J9M882 A0A1S3KDQ1 A0A151IKZ0 A0A0J7KCJ1 A0A2H9T450 A0A151JP94 X1XR94 A0A151I918 A0A2S2PJX4 J9L496 V9KQC4 A0A2S2P913 A0A2S2NDB3 A0A164G841 A0A151WZM5 A0A2S2PE96 A0A2S2QV10 A0A026X111 X1WN45 J9M6P6 A0A026W0I6 K7JK45 A0A151J7I0 A0A1X7VJ44 V5G7Y9 A0A1Y1M7F9 A0A2S2RBA8 A0A1Y1LRB1
EMBL
GGMR01018315
MBY30934.1
GGMR01008877
MBY21496.1
GFAA01003312
JAU00123.1
+ More
ABLF02009142 GGMS01007890 MBY77093.1 ABLF02014501 ABLF02029627 ABLF02007883 ABLF02066836 KQ977196 KYN05017.1 LBMM01009681 KMQ87949.1 NSIT01000305 PJE77982.1 KQ978756 KYN28602.1 ABLF02018482 KQ978319 KYM95153.1 GGMR01017131 MBY29750.1 ABLF02011475 JW868355 AFP00873.1 GGMR01013263 MBY25882.1 GGMR01002506 MBY15125.1 LRGB01016321 KZR98641.1 KQ982634 KYQ53351.1 GGMR01015158 MBY27777.1 GGMS01012375 MBY81578.1 KK107048 EZA61676.1 ABLF02008418 ABLF02042741 ABLF02013280 ABLF02042742 KK107512 EZA49485.1 AAZX01006011 KQ979690 KYN19755.1 GALX01002231 JAB66235.1 GEZM01038362 JAV81663.1 GGMS01017449 MBY86652.1 GEZM01049016 GEZM01049015 GEZM01049014 GEZM01049013 GEZM01049012 GEZM01049011 GEZM01049009 GEZM01049008 GEZM01049006 GEZM01049005 GEZM01049004 GEZM01049003 GEZM01049002 JAV76189.1
ABLF02009142 GGMS01007890 MBY77093.1 ABLF02014501 ABLF02029627 ABLF02007883 ABLF02066836 KQ977196 KYN05017.1 LBMM01009681 KMQ87949.1 NSIT01000305 PJE77982.1 KQ978756 KYN28602.1 ABLF02018482 KQ978319 KYM95153.1 GGMR01017131 MBY29750.1 ABLF02011475 JW868355 AFP00873.1 GGMR01013263 MBY25882.1 GGMR01002506 MBY15125.1 LRGB01016321 KZR98641.1 KQ982634 KYQ53351.1 GGMR01015158 MBY27777.1 GGMS01012375 MBY81578.1 KK107048 EZA61676.1 ABLF02008418 ABLF02042741 ABLF02013280 ABLF02042742 KK107512 EZA49485.1 AAZX01006011 KQ979690 KYN19755.1 GALX01002231 JAB66235.1 GEZM01038362 JAV81663.1 GGMS01017449 MBY86652.1 GEZM01049016 GEZM01049015 GEZM01049014 GEZM01049013 GEZM01049012 GEZM01049011 GEZM01049009 GEZM01049008 GEZM01049006 GEZM01049005 GEZM01049004 GEZM01049003 GEZM01049002 JAV76189.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A1W5BJ07
A0A2S2PNI2
A0A2S2NXS1
A0A1E1XL94
J9M9C6
A0A2S2QHC7
+ More
X1X3F6 J9KTJ5 J9M882 A0A1S3KDQ1 A0A151IKZ0 A0A0J7KCJ1 A0A2H9T450 A0A151JP94 X1XR94 A0A151I918 A0A2S2PJX4 J9L496 V9KQC4 A0A2S2P913 A0A2S2NDB3 A0A164G841 A0A151WZM5 A0A2S2PE96 A0A2S2QV10 A0A026X111 X1WN45 J9M6P6 A0A026W0I6 K7JK45 A0A151J7I0 A0A1X7VJ44 V5G7Y9 A0A1Y1M7F9 A0A2S2RBA8 A0A1Y1LRB1
X1X3F6 J9KTJ5 J9M882 A0A1S3KDQ1 A0A151IKZ0 A0A0J7KCJ1 A0A2H9T450 A0A151JP94 X1XR94 A0A151I918 A0A2S2PJX4 J9L496 V9KQC4 A0A2S2P913 A0A2S2NDB3 A0A164G841 A0A151WZM5 A0A2S2PE96 A0A2S2QV10 A0A026X111 X1WN45 J9M6P6 A0A026W0I6 K7JK45 A0A151J7I0 A0A1X7VJ44 V5G7Y9 A0A1Y1M7F9 A0A2S2RBA8 A0A1Y1LRB1
Ontologies
GO
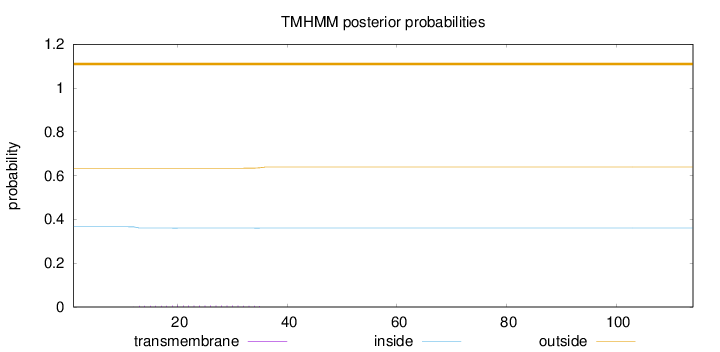
Topology
Length:
114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13186
Exp number, first 60 AAs:
0.13186
Total prob of N-in:
0.36625
outside
1 - 114
Population Genetic Test Statistics
Pi
321.619758
Theta
187.23879
Tajima's D
2.258468
CLR
0
CSRT
0.91930403479826
Interpretation
Uncertain
