Gene
KWMTBOMO05236
Annotation
PREDICTED:_tetracycline_resistance_protein?_class_B-like_isoform_X1_[Amyelois_transitella]
Full name
Major facilitator superfamily domain-containing protein 9
Location in the cell
PlasmaMembrane Reliability : 4.877
Sequence
CDS
ATGGCTACGCGAATATTGCAATCTATTAGTTTTTTGGATATGTTCGGAGCGGGCTTGTTGATGCCGCAACTAGCCAATCATGCGAAACAATTAGGATGCAGTCACATACAAATAGGAACGATGAGTGCTCTGTACTCTGGCTTCCAGCTGATATCCGGACCTGTCGTCGGAAGTCTCAGTGACACAAAAGGCAGGAAACCAATACTATTGTTGTGCCTGTACATCTGTGCATTCGCATATGTGTCGCTCGGGATTACATCCTCAATTACGCTATTTCTATTGTTAAGATCTGTGATAGGCATGTCCAAACAAACCCAACTATTGACGAACGCTTTAGCGCCAGACTATGAGACCGATGGGACCAAACAATCAATAGTATATGGAAGGATGTCGACAATGACTGGACTTGGAATGTCTGTGGGTCCTATCATTGGAGGACACATAATGGAAGCCTATCCCAATAACGGATTCACTGTAATTTGCTGTGTAGTCGGTGCTATATTCATTTTCAATACATTTTTGCTCAGAAAGCTGCCTGATGTAAAGATTTCGAAATTTGCCAAAAAACAAGTCACCATACCGATAAAACTTTTAGATTCAATTACAAAAACAGCTAAAGAATCCATTGATGAATTTCGCAAAATTGACTGGAGCATCTTCTGGGATTTGTTTTTATTTAAGCTCCTTATAACATGTTGTGTTGGATTGTACTTCAGTAGTTTTAGTTTATTTTTAAAAATATACCATGAGGTGTCTCCAAAAGAATTGGGATATTTAATAGCCTTTCAAGGCGGTATGGGATCTTTATGCACGTATTTCATTGGCCACATCACGAAATTGTACAAACACGATGAAGATTTTTCTCAAAGAATGTTGCATATTTCACTTGTATTATCTTTTACTTTATTGGGTTTATCCATGATTTCCAGTATCGTTATTTTTGTGTTGCTATTAATACCTTTTGCTTTATGTGGATCAATAGGAAGAGTTACCACTTTGGAAATGATTGCTAGTAGAAGTAAAAACACAAATAGAGGAACTATAATCGGTGCCTCAAATAGTATGAAATCTTTATCTGGGGTAATAACCCCATTGCTTGGTGGTTTTATTGGAGAGTATTTAGGTATAGCTTATAATTTCTACTTAGCCTCATTTGTGGCTACCGCTGGAGCAGTAGCTATAAAATATCATAGAACATACGTAACATCGAAAAATAAATCTGAATAA
Protein
MATRILQSISFLDMFGAGLLMPQLANHAKQLGCSHIQIGTMSALYSGFQLISGPVVGSLSDTKGRKPILLLCLYICAFAYVSLGITSSITLFLLLRSVIGMSKQTQLLTNALAPDYETDGTKQSIVYGRMSTMTGLGMSVGPIIGGHIMEAYPNNGFTVICCVVGAIFIFNTFLLRKLPDVKISKFAKKQVTIPIKLLDSITKTAKESIDEFRKIDWSIFWDLFLFKLLITCCVGLYFSSFSLFLKIYHEVSPKELGYLIAFQGGMGSLCTYFIGHITKLYKHDEDFSQRMLHISLVLSFTLLGLSMISSIVIFVLLLIPFALCGSIGRVTTLEMIASRSKNTNRGTIIGASNSMKSLSGVITPLLGGFIGEYLGIAYNFYLASFVATAGAVAIKYHRTYVTSKNKSE
Summary
Similarity
Belongs to the major facilitator superfamily.
Keywords
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Major facilitator superfamily domain-containing protein 9
Uniprot
A0A2H1VPF5
I4DKT2
A0A194PP29
A0A212FGB4
S4PDY8
H9IYV9
+ More
A0A2A4J545 A0A0L7KTY1 A0A0L7LSA0 A0A2A4J523 A0A2H1W5B2 A0A0T6B2F4 A0A1Y1N5D7 A0A1B6DT32 D6X4Q1 A0A0N7ZC16 A0A1Y1N5K2 A0A1W4XAB0 A0A2J7RN81 A0A1S3IUA9 A0A210PR03 A0A1B6FLP2 A0A1S3HPX7 A0A1B6DTI1 A0A3R7MFI1 K9LEA2 A0A1S3IUA4 A0A2S2QJE0 M3XHC1 A0A3Q3D4T5 A0A0P7TIV9 E9QH17 A0A1B6LC57 H3AR67 W5MQA0 A0A1B6MRL5 A0A0L8G7R9 A0A2B4RGX4 V4AMT5 A0A1B6HL99 W5MQ91 A0A3P8UWF7 Q8C0T7 R7UB65 V9KSU8 A0A1L8HGP7 A0A3B4GC69 W5KCK7 A0A3B4BTY2 A0A3Q3MPJ4 A0A1U7QBE6 A0A369SJQ3 A0A1U8DCW0 A0A0L8G7B0 J9JQX1 A0A2I0U5C2 B3RXQ7 A0A3Q2I9D2 A0A3Q0CZB7 A0A2J7Q663 A0A3Q2DT68 A0A2P8Z1R8 A0A1W4ZXI4 A0A1S3FGN7 A0A3Q3FRW8 G1NPN5 A0A210PR23 K1PPS4 A0A2S2P3K8 A0A099YUZ7 A0A3L7INK5 A0A3B3SF14 A0A1B6IHD6 A0A1V4J3P3 A0A1Y1JUC5 A0A2D0R4V1 A0A3Q2PFA8 G1PL12 A0A3M0K0G8 K9ITY8 H3D8P1 D3ZTY6 H2UDK3 A0A2P8Z1S0 A7RPG8 A0A3B4GDV8 A0A2G9S2L9 A0A093H1Y7 A0A3Q0ILX5 A0A3B4A6V6 A0A093PRR4 A0A1S3ITJ0 A0A1A8BKC9 A0A2I3S7B7 H2QIH0 A0A2R9BZN6
A0A2A4J545 A0A0L7KTY1 A0A0L7LSA0 A0A2A4J523 A0A2H1W5B2 A0A0T6B2F4 A0A1Y1N5D7 A0A1B6DT32 D6X4Q1 A0A0N7ZC16 A0A1Y1N5K2 A0A1W4XAB0 A0A2J7RN81 A0A1S3IUA9 A0A210PR03 A0A1B6FLP2 A0A1S3HPX7 A0A1B6DTI1 A0A3R7MFI1 K9LEA2 A0A1S3IUA4 A0A2S2QJE0 M3XHC1 A0A3Q3D4T5 A0A0P7TIV9 E9QH17 A0A1B6LC57 H3AR67 W5MQA0 A0A1B6MRL5 A0A0L8G7R9 A0A2B4RGX4 V4AMT5 A0A1B6HL99 W5MQ91 A0A3P8UWF7 Q8C0T7 R7UB65 V9KSU8 A0A1L8HGP7 A0A3B4GC69 W5KCK7 A0A3B4BTY2 A0A3Q3MPJ4 A0A1U7QBE6 A0A369SJQ3 A0A1U8DCW0 A0A0L8G7B0 J9JQX1 A0A2I0U5C2 B3RXQ7 A0A3Q2I9D2 A0A3Q0CZB7 A0A2J7Q663 A0A3Q2DT68 A0A2P8Z1R8 A0A1W4ZXI4 A0A1S3FGN7 A0A3Q3FRW8 G1NPN5 A0A210PR23 K1PPS4 A0A2S2P3K8 A0A099YUZ7 A0A3L7INK5 A0A3B3SF14 A0A1B6IHD6 A0A1V4J3P3 A0A1Y1JUC5 A0A2D0R4V1 A0A3Q2PFA8 G1PL12 A0A3M0K0G8 K9ITY8 H3D8P1 D3ZTY6 H2UDK3 A0A2P8Z1S0 A7RPG8 A0A3B4GDV8 A0A2G9S2L9 A0A093H1Y7 A0A3Q0ILX5 A0A3B4A6V6 A0A093PRR4 A0A1S3ITJ0 A0A1A8BKC9 A0A2I3S7B7 H2QIH0 A0A2R9BZN6
Pubmed
22651552
26354079
22118469
23622113
19121390
26227816
+ More
28004739 18362917 19820115 28812685 9215903 23594743 23254933 24487278 16141072 15489334 24402279 27762356 25329095 30042472 18719581 19892987 29403074 20838655 22992520 29704459 29240929 21993624 15496914 15057822 15632090 21551351 17615350 25463417 16136131 22722832
28004739 18362917 19820115 28812685 9215903 23594743 23254933 24487278 16141072 15489334 24402279 27762356 25329095 30042472 18719581 19892987 29403074 20838655 22992520 29704459 29240929 21993624 15496914 15057822 15632090 21551351 17615350 25463417 16136131 22722832
EMBL
ODYU01003479
SOQ42332.1
AK401900
BAM18522.1
KQ459597
KPI94738.1
+ More
AGBW02008688 OWR52776.1 GAIX01003436 JAA89124.1 BABH01010684 NWSH01003341 PCG66512.1 JTDY01005703 KOB66723.1 JTDY01000213 KOB78259.1 PCG66513.1 ODYU01006433 SOQ48271.1 LJIG01016136 KRT81506.1 GEZM01012179 JAV93132.1 GEDC01008483 JAS28815.1 KQ971380 EEZ97676.1 GDRN01074705 GDRN01074704 JAI63192.1 GEZM01012178 JAV93134.1 NEVH01002545 PNF42296.1 NEDP02005552 OWF38929.1 GECZ01018662 JAS51107.1 GEDC01008349 JAS28949.1 QCYY01001129 ROT80335.1 JQ278010 AFI81411.1 GGMS01008437 MBY77640.1 AFYH01107362 AFYH01107363 JARO02010263 KPP60834.1 CT030738 GEBQ01018701 JAT21276.1 AHAT01003416 GEBQ01001406 JAT38571.1 KQ423432 KOF72909.1 LSMT01000461 PFX17624.1 KB201721 ESO94911.1 GECU01032234 JAS75472.1 AK029874 BC057633 AMQN01001489 KB303020 ELU03605.1 JW868934 AFP01452.1 CM004468 OCT95270.1 NOWV01000006 RDD46732.1 KOF72912.1 ABLF02030142 KZ506143 PKU41235.1 DS985245 EDV24900.1 NEVH01017540 PNF24066.1 PYGN01000238 PSN50442.1 OWF38928.1 JH818841 EKC18435.1 GGMR01011391 MBY24010.1 KL885237 KGL72818.1 RAZU01000007 RLQ79580.1 GECU01021383 GECU01008020 JAS86323.1 JAS99686.1 LSYS01009367 OPJ66780.1 GEZM01102869 JAV51758.1 AAPE02041349 AAPE02041350 AAPE02041351 QRBI01000120 RMC06636.1 GABZ01001563 JAA51962.1 AABR07067464 CH473965 EDL99164.1 PSN50441.1 DS469525 EDO46735.1 KV929322 PIO34408.1 KL205917 KFV76598.1 KL670503 KFW79508.1 HADZ01003140 SBP67081.1 AACZ04055746 GABC01001273 GABC01001272 GABF01006514 GABF01006513 GABF01006512 GABF01006511 GABD01009663 GABE01011423 GABE01011422 GABE01011421 GABE01011420 NBAG03000391 JAA10065.1 JAA15631.1 JAA23437.1 JAA33316.1 PNI30442.1 AJFE02037131 AJFE02037132
AGBW02008688 OWR52776.1 GAIX01003436 JAA89124.1 BABH01010684 NWSH01003341 PCG66512.1 JTDY01005703 KOB66723.1 JTDY01000213 KOB78259.1 PCG66513.1 ODYU01006433 SOQ48271.1 LJIG01016136 KRT81506.1 GEZM01012179 JAV93132.1 GEDC01008483 JAS28815.1 KQ971380 EEZ97676.1 GDRN01074705 GDRN01074704 JAI63192.1 GEZM01012178 JAV93134.1 NEVH01002545 PNF42296.1 NEDP02005552 OWF38929.1 GECZ01018662 JAS51107.1 GEDC01008349 JAS28949.1 QCYY01001129 ROT80335.1 JQ278010 AFI81411.1 GGMS01008437 MBY77640.1 AFYH01107362 AFYH01107363 JARO02010263 KPP60834.1 CT030738 GEBQ01018701 JAT21276.1 AHAT01003416 GEBQ01001406 JAT38571.1 KQ423432 KOF72909.1 LSMT01000461 PFX17624.1 KB201721 ESO94911.1 GECU01032234 JAS75472.1 AK029874 BC057633 AMQN01001489 KB303020 ELU03605.1 JW868934 AFP01452.1 CM004468 OCT95270.1 NOWV01000006 RDD46732.1 KOF72912.1 ABLF02030142 KZ506143 PKU41235.1 DS985245 EDV24900.1 NEVH01017540 PNF24066.1 PYGN01000238 PSN50442.1 OWF38928.1 JH818841 EKC18435.1 GGMR01011391 MBY24010.1 KL885237 KGL72818.1 RAZU01000007 RLQ79580.1 GECU01021383 GECU01008020 JAS86323.1 JAS99686.1 LSYS01009367 OPJ66780.1 GEZM01102869 JAV51758.1 AAPE02041349 AAPE02041350 AAPE02041351 QRBI01000120 RMC06636.1 GABZ01001563 JAA51962.1 AABR07067464 CH473965 EDL99164.1 PSN50441.1 DS469525 EDO46735.1 KV929322 PIO34408.1 KL205917 KFV76598.1 KL670503 KFW79508.1 HADZ01003140 SBP67081.1 AACZ04055746 GABC01001273 GABC01001272 GABF01006514 GABF01006513 GABF01006512 GABF01006511 GABD01009663 GABE01011423 GABE01011422 GABE01011421 GABE01011420 NBAG03000391 JAA10065.1 JAA15631.1 JAA23437.1 JAA33316.1 PNI30442.1 AJFE02037131 AJFE02037132
Proteomes
UP000053268
UP000007151
UP000005204
UP000218220
UP000037510
UP000007266
+ More
UP000192223 UP000235965 UP000085678 UP000242188 UP000283509 UP000008672 UP000264820 UP000034805 UP000192224 UP000000437 UP000018468 UP000053454 UP000225706 UP000030746 UP000265120 UP000000589 UP000014760 UP000186698 UP000261460 UP000018467 UP000261440 UP000261640 UP000189706 UP000253843 UP000189705 UP000007819 UP000009022 UP000002281 UP000265020 UP000245037 UP000081671 UP000261660 UP000001645 UP000005408 UP000053641 UP000273346 UP000261540 UP000190648 UP000221080 UP000265000 UP000001074 UP000269221 UP000007303 UP000002494 UP000005226 UP000001593 UP000053584 UP000079169 UP000261520 UP000053258 UP000002277 UP000240080
UP000192223 UP000235965 UP000085678 UP000242188 UP000283509 UP000008672 UP000264820 UP000034805 UP000192224 UP000000437 UP000018468 UP000053454 UP000225706 UP000030746 UP000265120 UP000000589 UP000014760 UP000186698 UP000261460 UP000018467 UP000261440 UP000261640 UP000189706 UP000253843 UP000189705 UP000007819 UP000009022 UP000002281 UP000265020 UP000245037 UP000081671 UP000261660 UP000001645 UP000005408 UP000053641 UP000273346 UP000261540 UP000190648 UP000221080 UP000265000 UP000001074 UP000269221 UP000007303 UP000002494 UP000005226 UP000001593 UP000053584 UP000079169 UP000261520 UP000053258 UP000002277 UP000240080
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1VPF5
I4DKT2
A0A194PP29
A0A212FGB4
S4PDY8
H9IYV9
+ More
A0A2A4J545 A0A0L7KTY1 A0A0L7LSA0 A0A2A4J523 A0A2H1W5B2 A0A0T6B2F4 A0A1Y1N5D7 A0A1B6DT32 D6X4Q1 A0A0N7ZC16 A0A1Y1N5K2 A0A1W4XAB0 A0A2J7RN81 A0A1S3IUA9 A0A210PR03 A0A1B6FLP2 A0A1S3HPX7 A0A1B6DTI1 A0A3R7MFI1 K9LEA2 A0A1S3IUA4 A0A2S2QJE0 M3XHC1 A0A3Q3D4T5 A0A0P7TIV9 E9QH17 A0A1B6LC57 H3AR67 W5MQA0 A0A1B6MRL5 A0A0L8G7R9 A0A2B4RGX4 V4AMT5 A0A1B6HL99 W5MQ91 A0A3P8UWF7 Q8C0T7 R7UB65 V9KSU8 A0A1L8HGP7 A0A3B4GC69 W5KCK7 A0A3B4BTY2 A0A3Q3MPJ4 A0A1U7QBE6 A0A369SJQ3 A0A1U8DCW0 A0A0L8G7B0 J9JQX1 A0A2I0U5C2 B3RXQ7 A0A3Q2I9D2 A0A3Q0CZB7 A0A2J7Q663 A0A3Q2DT68 A0A2P8Z1R8 A0A1W4ZXI4 A0A1S3FGN7 A0A3Q3FRW8 G1NPN5 A0A210PR23 K1PPS4 A0A2S2P3K8 A0A099YUZ7 A0A3L7INK5 A0A3B3SF14 A0A1B6IHD6 A0A1V4J3P3 A0A1Y1JUC5 A0A2D0R4V1 A0A3Q2PFA8 G1PL12 A0A3M0K0G8 K9ITY8 H3D8P1 D3ZTY6 H2UDK3 A0A2P8Z1S0 A7RPG8 A0A3B4GDV8 A0A2G9S2L9 A0A093H1Y7 A0A3Q0ILX5 A0A3B4A6V6 A0A093PRR4 A0A1S3ITJ0 A0A1A8BKC9 A0A2I3S7B7 H2QIH0 A0A2R9BZN6
A0A2A4J545 A0A0L7KTY1 A0A0L7LSA0 A0A2A4J523 A0A2H1W5B2 A0A0T6B2F4 A0A1Y1N5D7 A0A1B6DT32 D6X4Q1 A0A0N7ZC16 A0A1Y1N5K2 A0A1W4XAB0 A0A2J7RN81 A0A1S3IUA9 A0A210PR03 A0A1B6FLP2 A0A1S3HPX7 A0A1B6DTI1 A0A3R7MFI1 K9LEA2 A0A1S3IUA4 A0A2S2QJE0 M3XHC1 A0A3Q3D4T5 A0A0P7TIV9 E9QH17 A0A1B6LC57 H3AR67 W5MQA0 A0A1B6MRL5 A0A0L8G7R9 A0A2B4RGX4 V4AMT5 A0A1B6HL99 W5MQ91 A0A3P8UWF7 Q8C0T7 R7UB65 V9KSU8 A0A1L8HGP7 A0A3B4GC69 W5KCK7 A0A3B4BTY2 A0A3Q3MPJ4 A0A1U7QBE6 A0A369SJQ3 A0A1U8DCW0 A0A0L8G7B0 J9JQX1 A0A2I0U5C2 B3RXQ7 A0A3Q2I9D2 A0A3Q0CZB7 A0A2J7Q663 A0A3Q2DT68 A0A2P8Z1R8 A0A1W4ZXI4 A0A1S3FGN7 A0A3Q3FRW8 G1NPN5 A0A210PR23 K1PPS4 A0A2S2P3K8 A0A099YUZ7 A0A3L7INK5 A0A3B3SF14 A0A1B6IHD6 A0A1V4J3P3 A0A1Y1JUC5 A0A2D0R4V1 A0A3Q2PFA8 G1PL12 A0A3M0K0G8 K9ITY8 H3D8P1 D3ZTY6 H2UDK3 A0A2P8Z1S0 A7RPG8 A0A3B4GDV8 A0A2G9S2L9 A0A093H1Y7 A0A3Q0ILX5 A0A3B4A6V6 A0A093PRR4 A0A1S3ITJ0 A0A1A8BKC9 A0A2I3S7B7 H2QIH0 A0A2R9BZN6
PDB
3WDO
E-value=0.0232724,
Score=88
Ontologies
KEGG
GO
Topology
Subcellular location
Membrane
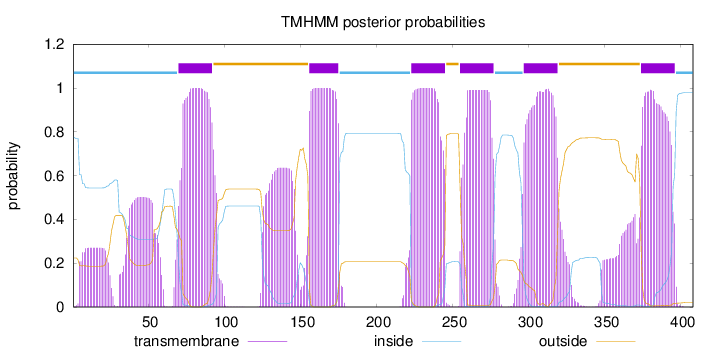
Length:
408
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
171.84969
Exp number, first 60 AAs:
16.58631
Total prob of N-in:
0.77629
POSSIBLE N-term signal
sequence
inside
1 - 69
TMhelix
70 - 92
outside
93 - 155
TMhelix
156 - 175
inside
176 - 222
TMhelix
223 - 245
outside
246 - 254
TMhelix
255 - 277
inside
278 - 296
TMhelix
297 - 319
outside
320 - 373
TMhelix
374 - 396
inside
397 - 408
Population Genetic Test Statistics
Pi
189.320919
Theta
167.380444
Tajima's D
0.481181
CLR
0.043963
CSRT
0.509974501274936
Interpretation
Uncertain
