Gene
KWMTBOMO05234
Pre Gene Modal
BGIBMGA002482
Annotation
PREDICTED:_DNA_excision_repair_protein_ERCC-1-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 2.746
Sequence
CDS
ATGGAAATAGAGGATGGTTTTGATGAATTGTTGGCCGAAATTAGTGAACCATCATCATCAAAGAAAATTAAAGTAGATGAAATTTCTGCTCAAGCAGGAACATCTGACGAAGCTACCATAAAACCAAGATCATCAAAGACACACTGTGTTCTAGTAAATAAAAACCAGAGAGGCAATCCTCTTCTCAAACACATAACCAGCGTGCCTTGGGAATATGATGATATAGTGCCTGATTATGAGGTTGGAAAAACAATATGCTTACTATTCCTCTCATTAAGATATCACAACTTGAATCCAGATTACATACACAACAGATTGAAGGAGCTAGGCAAGAAGTATGATTTGAGAGTTTTGCTAGTACAGGTTGATTTGAAAGATCCACATGCATCTTTAAAAAATTTAACACGAATATGCCTGCTGACTGATATAACTTTAATGTTAGCTTGGAACCCAGAGGAAGCAGCTAAAGTTGTTGAAAACTATAAAATTTACGAAAACAAACCGCCAGACAGAATTATGGAAAAAATTGAGAATGATCCTCATCAGAAGATTATCAATGCATTGTCGTCAATCAAACCTGTCAACAAAACTGATGCCATGACACTAATCAAAACATTTGGCACCCTAGAGAATATTATTAAAGTTTCAGAGTCTAGATTAGCAGAATGCCCTGGGTTTGGTATAACAAAAGCAAAAAAACTTTACAAGGCATTGCATGAACCATTTTTAAAGAAGGGTCAAACAAAAGATAAAAAAGATGAATTTCCTGATGAAGATTTGACACTTGAAGAATTAGAGAAAATTGTAAATGAAAACCAAGATATATCCTGA
Protein
MEIEDGFDELLAEISEPSSSKKIKVDEISAQAGTSDEATIKPRSSKTHCVLVNKNQRGNPLLKHITSVPWEYDDIVPDYEVGKTICLLFLSLRYHNLNPDYIHNRLKELGKKYDLRVLLVQVDLKDPHASLKNLTRICLLTDITLMLAWNPEEAAKVVENYKIYENKPPDRIMEKIENDPHQKIINALSSIKPVNKTDAMTLIKTFGTLENIIKVSESRLAECPGFGITKAKKLYKALHEPFLKKGQTKDKKDEFPDEDLTLEELEKIVNENQDIS
Summary
Uniprot
H9IYZ7
A0A2H1W5I2
A0A212FPC9
S4PXW7
A0A194QHF8
A0A0L7LFA1
+ More
A0A023ELD2 Q16ZS1 Q16IF3 A0A182GD33 A0A1B6J004 A0A1B6EQ02 A0A1Q3F5R7 A0A232FCR2 A0A1B0DB84 K7J2B8 A0A336M8L9 A0A2Z5TRG1 B0X6V4 A0A182K5H1 A0A182I869 A0A182KML6 A0A182XKI1 A0A2R7WFB7 A0A182NDX9 A0A182J1J9 A0A182UYE4 A0A182UCX8 Q7Q256 A0A1B6BZS6 A0A084VTL1 A0A182MJC1 A0A2J7RN28 A0A182P430 A0A0L7QLF1 A0A1B0FBW3 B3MC73 A0A182YG11 A0A1J1IWY4 A0A1B6M6T3 A0A1B0A0B4 A0A182QC63 A0A1Y1KIN0 A0A182RFJ8 J3JUT2 B4LJT8 A0A182VWZ6 Q292E9 A0A171AGE1 B4GDM6 A0A182T3H3 A0A087ZQZ8 A0A2M4BZ61 A0A2A3E192 A0A0P4VU68 Q7KMG7 A0A2H8TK95 A0A1B0BJL5 A0A0C9R016 A0A224XW85 A0A1A9YQ96 A0A182FA36 A0A154PT00 R4G7V1 B4JW82 A0A0L0C8T0 B4MY76 A0A3B0JTI0 J9JUK8 A0A310STA4 A0A0J9RCR8 A0A2M4BY99 B4QFZ7 Q95S37 A0A069DRD6 A0A0N0BF57 U5ER47 B4HRQ5 B4KT66 A0A2S2N6H8 B4P7Q2 A0A2S2R4W7 A0A1W4U640 A0A1I8MQQ3 A0A0M4EH38 W5JH12 A0A1A9ULN9 A0A0T6B1Z5 B3NQR6 A0A1I8NU01 A0A195CJZ1 A0A1S3JP97 A0A151XAL8 A0A0A9Y731 A0A195B763 A0A146MCN6 A0A195FG53 A0A146KPP7
A0A023ELD2 Q16ZS1 Q16IF3 A0A182GD33 A0A1B6J004 A0A1B6EQ02 A0A1Q3F5R7 A0A232FCR2 A0A1B0DB84 K7J2B8 A0A336M8L9 A0A2Z5TRG1 B0X6V4 A0A182K5H1 A0A182I869 A0A182KML6 A0A182XKI1 A0A2R7WFB7 A0A182NDX9 A0A182J1J9 A0A182UYE4 A0A182UCX8 Q7Q256 A0A1B6BZS6 A0A084VTL1 A0A182MJC1 A0A2J7RN28 A0A182P430 A0A0L7QLF1 A0A1B0FBW3 B3MC73 A0A182YG11 A0A1J1IWY4 A0A1B6M6T3 A0A1B0A0B4 A0A182QC63 A0A1Y1KIN0 A0A182RFJ8 J3JUT2 B4LJT8 A0A182VWZ6 Q292E9 A0A171AGE1 B4GDM6 A0A182T3H3 A0A087ZQZ8 A0A2M4BZ61 A0A2A3E192 A0A0P4VU68 Q7KMG7 A0A2H8TK95 A0A1B0BJL5 A0A0C9R016 A0A224XW85 A0A1A9YQ96 A0A182FA36 A0A154PT00 R4G7V1 B4JW82 A0A0L0C8T0 B4MY76 A0A3B0JTI0 J9JUK8 A0A310STA4 A0A0J9RCR8 A0A2M4BY99 B4QFZ7 Q95S37 A0A069DRD6 A0A0N0BF57 U5ER47 B4HRQ5 B4KT66 A0A2S2N6H8 B4P7Q2 A0A2S2R4W7 A0A1W4U640 A0A1I8MQQ3 A0A0M4EH38 W5JH12 A0A1A9ULN9 A0A0T6B1Z5 B3NQR6 A0A1I8NU01 A0A195CJZ1 A0A1S3JP97 A0A151XAL8 A0A0A9Y731 A0A195B763 A0A146MCN6 A0A195FG53 A0A146KPP7
Pubmed
19121390
22118469
23622113
26354079
26227816
24945155
+ More
17510324 26483478 28648823 20075255 26760975 20966253 12364791 14747013 17210077 24438588 17994087 25244985 28004739 22516182 23537049 15632085 27129103 10812334 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 22936249 26334808 17550304 25315136 20920257 23761445 25401762 26823975
17510324 26483478 28648823 20075255 26760975 20966253 12364791 14747013 17210077 24438588 17994087 25244985 28004739 22516182 23537049 15632085 27129103 10812334 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 22936249 26334808 17550304 25315136 20920257 23761445 25401762 26823975
EMBL
BABH01010810
ODYU01006433
SOQ48273.1
AGBW02003014
OWR55585.1
GAIX01004318
+ More
JAA88242.1 KQ459220 KPJ02876.1 JTDY01001358 KOB74085.1 GAPW01003560 JAC10038.1 CH477485 EAT40150.1 CH478081 EAT34045.1 JXUM01160686 KQ573783 KXJ67905.1 GECU01015217 JAS92489.1 GECZ01029754 JAS40015.1 GFDL01012130 JAV22915.1 NNAY01000456 OXU28270.1 AJVK01004674 AAZX01001561 UFQS01000673 UFQT01000673 SSX06033.1 SSX26390.1 FX985813 BBA93700.1 DS232425 EDS41625.1 APCN01002659 KK854737 PTY18306.1 AAAB01008978 EAA13582.4 GEDC01030823 JAS06475.1 ATLV01016361 KE525079 KFB41305.1 AXCM01002744 NEVH01002545 PNF42236.1 KQ414915 KOC59443.1 CCAG010009805 CH902619 EDV36173.1 CVRI01000063 CRL04717.1 GEBQ01008342 JAT31635.1 AXCN02002211 GEZM01086418 GEZM01086417 JAV59465.1 APGK01056773 BT126997 KB741277 KB632196 AEE61959.1 ENN71111.1 ERL89972.1 CH940648 EDW60597.1 CM000071 EAL24913.1 GEMB01001097 JAS02050.1 CH479181 EDW31683.1 GGFJ01008897 MBW58038.1 KZ288474 PBC25294.1 GDKW01002226 JAI54369.1 AF146797 AE013599 AAD38010.1 AAF58196.1 GFXV01001903 MBW13708.1 JXJN01015446 GBYB01001295 JAG71062.1 GFTR01004157 JAW12269.1 KQ435180 KZC14993.1 ACPB03005165 GAHY01002082 JAA75428.1 CH916375 EDV98220.1 JRES01000755 KNC28642.1 CH963894 EDW77065.1 OUUW01000001 SPP74378.1 ABLF02026344 ABLF02026345 KQ760176 OAD61645.1 CM002911 KMY93853.1 GGFJ01008898 MBW58039.1 CM000362 EDX07142.1 AY060971 AAL28519.1 GBGD01002628 JAC86261.1 KQ435812 KOX72862.1 GANO01003933 JAB55938.1 CH480816 EDW47919.1 CH933808 EDW09586.1 GGMR01000146 MBY12765.1 CM000158 EDW91078.1 KRJ99599.1 GGMS01015760 MBY84963.1 CP012524 ALC41822.1 ADMH02001261 ETN63351.1 LJIG01016172 KRT81400.1 CH954179 EDV55976.1 KQ977721 KYN00389.1 KQ982339 KYQ57397.1 GBHO01015637 GBHO01015635 GBRD01008148 JAG27967.1 JAG27969.1 JAG57673.1 KQ976574 KYM80358.1 GDHC01002319 JAQ16310.1 KQ981625 KYN39004.1 GDHC01021639 JAP96989.1
JAA88242.1 KQ459220 KPJ02876.1 JTDY01001358 KOB74085.1 GAPW01003560 JAC10038.1 CH477485 EAT40150.1 CH478081 EAT34045.1 JXUM01160686 KQ573783 KXJ67905.1 GECU01015217 JAS92489.1 GECZ01029754 JAS40015.1 GFDL01012130 JAV22915.1 NNAY01000456 OXU28270.1 AJVK01004674 AAZX01001561 UFQS01000673 UFQT01000673 SSX06033.1 SSX26390.1 FX985813 BBA93700.1 DS232425 EDS41625.1 APCN01002659 KK854737 PTY18306.1 AAAB01008978 EAA13582.4 GEDC01030823 JAS06475.1 ATLV01016361 KE525079 KFB41305.1 AXCM01002744 NEVH01002545 PNF42236.1 KQ414915 KOC59443.1 CCAG010009805 CH902619 EDV36173.1 CVRI01000063 CRL04717.1 GEBQ01008342 JAT31635.1 AXCN02002211 GEZM01086418 GEZM01086417 JAV59465.1 APGK01056773 BT126997 KB741277 KB632196 AEE61959.1 ENN71111.1 ERL89972.1 CH940648 EDW60597.1 CM000071 EAL24913.1 GEMB01001097 JAS02050.1 CH479181 EDW31683.1 GGFJ01008897 MBW58038.1 KZ288474 PBC25294.1 GDKW01002226 JAI54369.1 AF146797 AE013599 AAD38010.1 AAF58196.1 GFXV01001903 MBW13708.1 JXJN01015446 GBYB01001295 JAG71062.1 GFTR01004157 JAW12269.1 KQ435180 KZC14993.1 ACPB03005165 GAHY01002082 JAA75428.1 CH916375 EDV98220.1 JRES01000755 KNC28642.1 CH963894 EDW77065.1 OUUW01000001 SPP74378.1 ABLF02026344 ABLF02026345 KQ760176 OAD61645.1 CM002911 KMY93853.1 GGFJ01008898 MBW58039.1 CM000362 EDX07142.1 AY060971 AAL28519.1 GBGD01002628 JAC86261.1 KQ435812 KOX72862.1 GANO01003933 JAB55938.1 CH480816 EDW47919.1 CH933808 EDW09586.1 GGMR01000146 MBY12765.1 CM000158 EDW91078.1 KRJ99599.1 GGMS01015760 MBY84963.1 CP012524 ALC41822.1 ADMH02001261 ETN63351.1 LJIG01016172 KRT81400.1 CH954179 EDV55976.1 KQ977721 KYN00389.1 KQ982339 KYQ57397.1 GBHO01015637 GBHO01015635 GBRD01008148 JAG27967.1 JAG27969.1 JAG57673.1 KQ976574 KYM80358.1 GDHC01002319 JAQ16310.1 KQ981625 KYN39004.1 GDHC01021639 JAP96989.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000008820
UP000069940
+ More
UP000249989 UP000215335 UP000092462 UP000002358 UP000002320 UP000075881 UP000075840 UP000075882 UP000076407 UP000075884 UP000075880 UP000075903 UP000075902 UP000007062 UP000030765 UP000075883 UP000235965 UP000075885 UP000053825 UP000092444 UP000007801 UP000076408 UP000183832 UP000092445 UP000075886 UP000075900 UP000019118 UP000030742 UP000008792 UP000075920 UP000001819 UP000008744 UP000075901 UP000005203 UP000242457 UP000000803 UP000092460 UP000092443 UP000069272 UP000076502 UP000015103 UP000001070 UP000037069 UP000007798 UP000268350 UP000007819 UP000000304 UP000053105 UP000001292 UP000009192 UP000002282 UP000192221 UP000095301 UP000092553 UP000000673 UP000078200 UP000008711 UP000095300 UP000078542 UP000085678 UP000075809 UP000078540 UP000078541
UP000249989 UP000215335 UP000092462 UP000002358 UP000002320 UP000075881 UP000075840 UP000075882 UP000076407 UP000075884 UP000075880 UP000075903 UP000075902 UP000007062 UP000030765 UP000075883 UP000235965 UP000075885 UP000053825 UP000092444 UP000007801 UP000076408 UP000183832 UP000092445 UP000075886 UP000075900 UP000019118 UP000030742 UP000008792 UP000075920 UP000001819 UP000008744 UP000075901 UP000005203 UP000242457 UP000000803 UP000092460 UP000092443 UP000069272 UP000076502 UP000015103 UP000001070 UP000037069 UP000007798 UP000268350 UP000007819 UP000000304 UP000053105 UP000001292 UP000009192 UP000002282 UP000192221 UP000095301 UP000092553 UP000000673 UP000078200 UP000008711 UP000095300 UP000078542 UP000085678 UP000075809 UP000078540 UP000078541
Interpro
ProteinModelPortal
H9IYZ7
A0A2H1W5I2
A0A212FPC9
S4PXW7
A0A194QHF8
A0A0L7LFA1
+ More
A0A023ELD2 Q16ZS1 Q16IF3 A0A182GD33 A0A1B6J004 A0A1B6EQ02 A0A1Q3F5R7 A0A232FCR2 A0A1B0DB84 K7J2B8 A0A336M8L9 A0A2Z5TRG1 B0X6V4 A0A182K5H1 A0A182I869 A0A182KML6 A0A182XKI1 A0A2R7WFB7 A0A182NDX9 A0A182J1J9 A0A182UYE4 A0A182UCX8 Q7Q256 A0A1B6BZS6 A0A084VTL1 A0A182MJC1 A0A2J7RN28 A0A182P430 A0A0L7QLF1 A0A1B0FBW3 B3MC73 A0A182YG11 A0A1J1IWY4 A0A1B6M6T3 A0A1B0A0B4 A0A182QC63 A0A1Y1KIN0 A0A182RFJ8 J3JUT2 B4LJT8 A0A182VWZ6 Q292E9 A0A171AGE1 B4GDM6 A0A182T3H3 A0A087ZQZ8 A0A2M4BZ61 A0A2A3E192 A0A0P4VU68 Q7KMG7 A0A2H8TK95 A0A1B0BJL5 A0A0C9R016 A0A224XW85 A0A1A9YQ96 A0A182FA36 A0A154PT00 R4G7V1 B4JW82 A0A0L0C8T0 B4MY76 A0A3B0JTI0 J9JUK8 A0A310STA4 A0A0J9RCR8 A0A2M4BY99 B4QFZ7 Q95S37 A0A069DRD6 A0A0N0BF57 U5ER47 B4HRQ5 B4KT66 A0A2S2N6H8 B4P7Q2 A0A2S2R4W7 A0A1W4U640 A0A1I8MQQ3 A0A0M4EH38 W5JH12 A0A1A9ULN9 A0A0T6B1Z5 B3NQR6 A0A1I8NU01 A0A195CJZ1 A0A1S3JP97 A0A151XAL8 A0A0A9Y731 A0A195B763 A0A146MCN6 A0A195FG53 A0A146KPP7
A0A023ELD2 Q16ZS1 Q16IF3 A0A182GD33 A0A1B6J004 A0A1B6EQ02 A0A1Q3F5R7 A0A232FCR2 A0A1B0DB84 K7J2B8 A0A336M8L9 A0A2Z5TRG1 B0X6V4 A0A182K5H1 A0A182I869 A0A182KML6 A0A182XKI1 A0A2R7WFB7 A0A182NDX9 A0A182J1J9 A0A182UYE4 A0A182UCX8 Q7Q256 A0A1B6BZS6 A0A084VTL1 A0A182MJC1 A0A2J7RN28 A0A182P430 A0A0L7QLF1 A0A1B0FBW3 B3MC73 A0A182YG11 A0A1J1IWY4 A0A1B6M6T3 A0A1B0A0B4 A0A182QC63 A0A1Y1KIN0 A0A182RFJ8 J3JUT2 B4LJT8 A0A182VWZ6 Q292E9 A0A171AGE1 B4GDM6 A0A182T3H3 A0A087ZQZ8 A0A2M4BZ61 A0A2A3E192 A0A0P4VU68 Q7KMG7 A0A2H8TK95 A0A1B0BJL5 A0A0C9R016 A0A224XW85 A0A1A9YQ96 A0A182FA36 A0A154PT00 R4G7V1 B4JW82 A0A0L0C8T0 B4MY76 A0A3B0JTI0 J9JUK8 A0A310STA4 A0A0J9RCR8 A0A2M4BY99 B4QFZ7 Q95S37 A0A069DRD6 A0A0N0BF57 U5ER47 B4HRQ5 B4KT66 A0A2S2N6H8 B4P7Q2 A0A2S2R4W7 A0A1W4U640 A0A1I8MQQ3 A0A0M4EH38 W5JH12 A0A1A9ULN9 A0A0T6B1Z5 B3NQR6 A0A1I8NU01 A0A195CJZ1 A0A1S3JP97 A0A151XAL8 A0A0A9Y731 A0A195B763 A0A146MCN6 A0A195FG53 A0A146KPP7
PDB
2A1I
E-value=1.95811e-45,
Score=458
Ontologies
KEGG
PATHWAY
GO
PANTHER
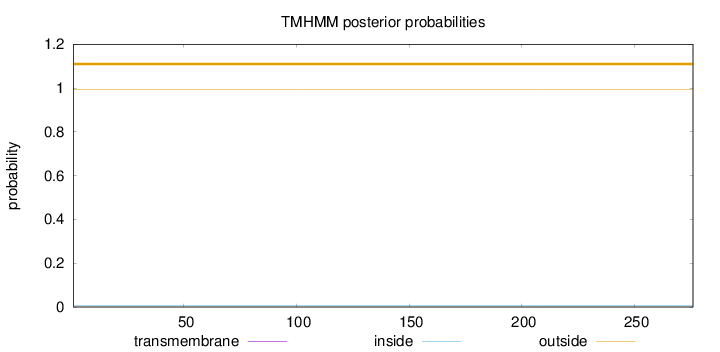
Topology
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00209
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00667
outside
1 - 276
Population Genetic Test Statistics
Pi
305.634237
Theta
223.648376
Tajima's D
1.223312
CLR
0
CSRT
0.718314084295785
Interpretation
Possibly Positive selection
