Pre Gene Modal
BGIBMGA002428
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_membrane-associated_protein_Hem_[Bombyx_mori]
Full name
Membrane-associated protein Hem
Alternative Name
dHem-2
Location in the cell
Cytoplasmic Reliability : 1.037 Nuclear Reliability : 1.55 PlasmaMembrane Reliability : 1.307
Sequence
CDS
ATGTCCACATTATCAAGATCCCTGAATATTAGTCAGCAAAAATTAGCTGAGAAGCTAATTATACTGAATGACAGAGGTATTGGTATTTTAACAAGAGTTTACAACATTAAGAAAGCCTGTGGAGATGCCAAATCGAAACCAGCATTTCTATCTGATAAAGCATTGGAGTCATCAATTAAGCACATAGTTAGGAGATTTCCCAACATTGACGTTAAAGGACTCCAAGCAATTACACCAATACGGAATGAAGTAATCAAATCATTGTCGCTATACTATTACACATTTGTAGATTTGTTAGATTTTAAAGACAATGTTTGTGAACTTCTTAACATCATAGATGCATGTCAGGTTACTTTAGACTTGACATTGAATTTTGAATTGACAAAAAACTATCTTGACTTGGTTTTAACATATGTTACACTAATGATATTGTTGTCTCGAGTTGAAGACAGAAAAGCAGTGTTAGGTTTATTTAATGCTGCATATGAATTGCTACAGAATCAAATTGAGAATAGTTTTCCTCGTTTGGGACAGTTGATTGTTGATTATGATGCTCCATTGAAGAAATTGTCTGAAGAATTTGTGCCACATCAAAAAGTACTAGCAAGTGCATTAAATTCTCTATGGCATGTGTATCCAGCTAGAAATCTTTCTGCTGAACATTGGAGATCAGAGCAAAAGCTAAGCTTAGTAAGTAATCCAGCTCAGTTACTGAAACCATCAGAAACAAACACAATGTCTTGTGAATATGTATCATTGGAGTCTTTGGAGAGGTGGGTCATATTTGGTTTTGCACTCTGTCATCAGATGCTCCAGCAGGACCATGCTAATAAAATGTGGGTCAATGCACTAGAGTCAGGATGGGTTGTGGCATTATTTAGAGATGAAGTGATTTACATTCACAGCTATATACAAGGTTTCTTTGACAGCATAAAAGGCTATGGCAAAAGAATATCTGAAGTAAAAGATTGTTACCATCATGCAGTACAGAAAGCAGGATACAAACACAGAGAGAGGAGAAAATTCTTAAGAACAGCTTTGAAAGAACTGGGTCTAATATTGACTGATCAACCAGGATTATTGGGCCCAAAAGCTCTTTTAATTTTCATTGGCCTCTGCTATGCAAGAGATGAAGTATTTTGGCTGTTGCGACATAATGATAACCCACCACAAAAAGTAAAGGGAAAATCTGCAGAAGATTTGGTAGACCGTCAACTTCCTGAACTTTTGTTCCACATGGAAGAATTAAGAGCATTGGTTCGTAAATATAGTCAGGTAATGCAGAGATACTATGTGCAATACCTTTCAGGCTTTGATGCTGTAGCATTGAACATGATGATTCAGAACTTGAAGGTATGTCCTGAAGACGAAAGTATAATATTATCATCTTTATGTACTACTGCAGCAAATTTGAGTGTCAAACAGGTTGAAGAGAATGAGCCATTTGATTTTCGATCTTTCCGTTTAGATTGGTTCAGACTTCAAGCATATACATCTGTTGTCAAAACCCCCTTGAGTCTTGTTGATCAAAGGGAACTGGCTCAGTTTATAGACAAAATGGTATTCCATACTAAAATGGTAGATAACCTTGATGAAATAATGGTTGAAACATCTGATCTGTCAATATTTTGCTTTTACAACAAAATATTTGAAAATCAGTTCCATATGTGTCTTGAGTTCCCTGCACAAAATCGGTATATTATAGCATTCCCGCTTATTTGTAGTCATTTCCAAAATTGTACTCATGAACTATGCCCTGAAGAGAGGCATCACATTCTAGAGAGAAGTTTGTCAGTAGTCAACATATTCTTGGATGAAATGGCAAAAGAAGCAAAAAACATTATCACCACTATATGTGATGAACAGTGTACTATGAGTGACAAATTACTACCCAAACATTGTGCACAAACTATAGCTCATTTAGCTAACAGAAAAAAGAAGGATAAAAATAAGAAAAACACAATTGAAATTGTTAAGCCTGGAGCAGAAAGTTACAGAAAAACCAGAGAAGAATTGACAACCATGGACAAACTTCACATGGCTTTGACAGAATTATGCTATGCTATAAATTATTGTTCCACTGTAAATGTTTGGGAGTATACTTTTGCACCACGAGAGTACTTGCACCAACATTTGGAAACTAGATTCTCTAAGGCCTTGGTTGGCATGGTAATGTTTAACCAAGACACAAGTGAAATTGCTAAGCCTTCTGAGTTGCTTGTTAGTGTTCGTGCATACATGAATGTGCTTCAAACTGTTGAAAATTACGTTCATATTGATATTACACGAGTTTTCAATAATTGTCTTTTACAACAAACTCAAAATATGGATAGTCATAAAGAAAAAACCATTGCATCATTGTATACACAGTGGTACTCTGAGATTTTACTGAGACGGGTCAGTGCTGGTAGTATTTGTTTTTCAATGAGTCAAAAAGCATTTGTTAGTTTGACAGCCGAAGGTGCTATACCCTTTAATGCCGAGGAATATTCTGATATTAATGAGTTAAGGGCATTAGCTGAATTAATTGGGCCCTATGGCATGAAACTGTTAAGTGAAACACTAATGTGGCAAATTGCAAGTCAAGTACAGGAATTGAAGAAACTGGTTGTACAAAATAAAGAGGTGTTGCAGATGCTCAGGACAAATTTTGATAAACCAGAAATTATGAGAGAGCAGTTCAAAAGATTGCAAAATGTGGATAATGTACTACAACGGATGACCATTATTGGAGTCATATTGAGTTTCAGACAAATAGCTCAAGAGTCATTACTAGATGTTTTAGAAAGGCGTATTCCGTTTTTGATAAGTTCCATAAAAGACTTCCAGCAACAGCTGCCGAGTGGTGATCCAATGCGAGTCATTTCAGAAATGTGTTCTGCTGCTGGCCTACCGTGCAAAGTGGATCCAACATTGGCATCAGCTTTACGTCAGCAAAAATCAGAAGTTGAGGAGGAAGAGCATTTAATTGTATGCTTGCTAATGGTGTTTGTTGCTGTGTCTTTGCCTAGACTAGCTCGGAGTGAAGTATCTTTCTATCGACCTTCCCTGGAAGGTCATGCTAACAATATACACTGTATAGCTCCAGCTATCAACCACATATTCGGTGCTTTATTCACTATATGCGGCCAGGGCGATATCGAGGATCGAATGAAAGAATTCTTAGCTTTAGCGTCATCATCTTTGCTTCGTTTAGGGCAAGAAACTGATAAAGAAGCTACTAAGAATAGGGAATCAGTTTACCTTCTATTAGACTTGATAGTGCAGGAGTCACCCTTTCTTACAATGGATTTGTTGGAGTCATGTTTTCCATATGTATTGATACGTAATGCTTATCATGAAGTTTATAAACAAGAGCAGATGCTTCTGCATATGTAA
Protein
MSTLSRSLNISQQKLAEKLIILNDRGIGILTRVYNIKKACGDAKSKPAFLSDKALESSIKHIVRRFPNIDVKGLQAITPIRNEVIKSLSLYYYTFVDLLDFKDNVCELLNIIDACQVTLDLTLNFELTKNYLDLVLTYVTLMILLSRVEDRKAVLGLFNAAYELLQNQIENSFPRLGQLIVDYDAPLKKLSEEFVPHQKVLASALNSLWHVYPARNLSAEHWRSEQKLSLVSNPAQLLKPSETNTMSCEYVSLESLERWVIFGFALCHQMLQQDHANKMWVNALESGWVVALFRDEVIYIHSYIQGFFDSIKGYGKRISEVKDCYHHAVQKAGYKHRERRKFLRTALKELGLILTDQPGLLGPKALLIFIGLCYARDEVFWLLRHNDNPPQKVKGKSAEDLVDRQLPELLFHMEELRALVRKYSQVMQRYYVQYLSGFDAVALNMMIQNLKVCPEDESIILSSLCTTAANLSVKQVEENEPFDFRSFRLDWFRLQAYTSVVKTPLSLVDQRELAQFIDKMVFHTKMVDNLDEIMVETSDLSIFCFYNKIFENQFHMCLEFPAQNRYIIAFPLICSHFQNCTHELCPEERHHILERSLSVVNIFLDEMAKEAKNIITTICDEQCTMSDKLLPKHCAQTIAHLANRKKKDKNKKNTIEIVKPGAESYRKTREELTTMDKLHMALTELCYAINYCSTVNVWEYTFAPREYLHQHLETRFSKALVGMVMFNQDTSEIAKPSELLVSVRAYMNVLQTVENYVHIDITRVFNNCLLQQTQNMDSHKEKTIASLYTQWYSEILLRRVSAGSICFSMSQKAFVSLTAEGAIPFNAEEYSDINELRALAELIGPYGMKLLSETLMWQIASQVQELKKLVVQNKEVLQMLRTNFDKPEIMREQFKRLQNVDNVLQRMTIIGVILSFRQIAQESLLDVLERRIPFLISSIKDFQQQLPSGDPMRVISEMCSAAGLPCKVDPTLASALRQQKSEVEEEEHLIVCLLMVFVAVSLPRLARSEVSFYRPSLEGHANNIHCIAPAINHIFGALFTICGQGDIEDRMKEFLALASSSLLRLGQETDKEATKNRESVYLLLDLIVQESPFLTMDLLESCFPYVLIRNAYHEVYKQEQMLLHM
Summary
Description
Plays a role during growth of the oocyte.
Subunit
Component of the WAVE complex composed of Hem/Kette, Scar/Wave and Sra-1/Cyfip where it binds directly to the C-terminus of Sra-1.
Similarity
Belongs to the heat shock protein 70 family.
Belongs to the HEM-1/HEM-2 family.
Belongs to the HEM-1/HEM-2 family.
Keywords
Cell membrane
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Membrane-associated protein Hem
Uniprot
A0A1E1VYL1
A0A194QBF4
A0A2H1VZQ4
A0A194RME1
A0A212FPB6
A0A0L7KR32
+ More
A0A1B6E8D2 A0A2J7RSR7 A0A067R1Z2 A0A0A9YNG5 A0A0C9RHN7 A0A151WUN7 A0A151JZS4 E2C9I5 F4X615 A0A195C7Q6 A0A151IY57 A0A0J7L6I5 E2ALH9 A0A151I1Z4 A0A158NMS9 E9J2G9 A0A2A3E3X8 K7IXQ7 A0A310SQH9 A0A1Y1K2U0 A0A026W2C6 A0A088AFJ6 A0A1W4WP41 A0A154PMQ3 E0W2A8 A0A023F447 A0A0L7QJK1 A0A0V0G7U7 A0A0T6BES9 Q16V64 A0A069DZQ9 A0A1Q3FXH3 A0A0N7Z970 A0A182H761 A0A2C9GQZ7 A0A182TK65 A0A182KNQ8 Q7PPI1 A0A182VI02 A0A182M787 A0A2M4AEB2 A0A182SLJ9 A0A182X2H7 W5J5D3 A0A182QSA6 A0A336LPG5 A0A182YKR9 A0A182WM09 A0A182JBE8 A0A182K4N4 A0A182N1Z7 A0A182P5G5 D7EI65 A0A182RAL7 W8AWC4 A0A0K8UG50 A0A034VHD8 A0A0A1XHY9 B4MND0 A0A0M4F0H1 B3MB43 A0A1A9WSP3 B4KX53 A0A1B0FKC3 A0A1A9XGF2 A0A1B0BZM6 A0A1A9ZKP0 A0A1A9UFU4 P55162 A0A1I8M685 B3NJ37 T1PFW3 A0A3B0JRM4 Q29E13 B4H430 A0A1W4UZ08 B4QKR9 B4LFH9 B4IAZ8 A0A0L0BUH4 B4J304 A0A1J1IGR0 A0A1B6KH25 A0A1I8Q3C5 A0A1J1J7Q4 H9IYU3 U4UE05 N6TSI2 A0A0P5RNZ5 E9FST5 A0A0P5XNV6 A0A0P5LU54 T1JIM6 A0A3R7M9U9
A0A1B6E8D2 A0A2J7RSR7 A0A067R1Z2 A0A0A9YNG5 A0A0C9RHN7 A0A151WUN7 A0A151JZS4 E2C9I5 F4X615 A0A195C7Q6 A0A151IY57 A0A0J7L6I5 E2ALH9 A0A151I1Z4 A0A158NMS9 E9J2G9 A0A2A3E3X8 K7IXQ7 A0A310SQH9 A0A1Y1K2U0 A0A026W2C6 A0A088AFJ6 A0A1W4WP41 A0A154PMQ3 E0W2A8 A0A023F447 A0A0L7QJK1 A0A0V0G7U7 A0A0T6BES9 Q16V64 A0A069DZQ9 A0A1Q3FXH3 A0A0N7Z970 A0A182H761 A0A2C9GQZ7 A0A182TK65 A0A182KNQ8 Q7PPI1 A0A182VI02 A0A182M787 A0A2M4AEB2 A0A182SLJ9 A0A182X2H7 W5J5D3 A0A182QSA6 A0A336LPG5 A0A182YKR9 A0A182WM09 A0A182JBE8 A0A182K4N4 A0A182N1Z7 A0A182P5G5 D7EI65 A0A182RAL7 W8AWC4 A0A0K8UG50 A0A034VHD8 A0A0A1XHY9 B4MND0 A0A0M4F0H1 B3MB43 A0A1A9WSP3 B4KX53 A0A1B0FKC3 A0A1A9XGF2 A0A1B0BZM6 A0A1A9ZKP0 A0A1A9UFU4 P55162 A0A1I8M685 B3NJ37 T1PFW3 A0A3B0JRM4 Q29E13 B4H430 A0A1W4UZ08 B4QKR9 B4LFH9 B4IAZ8 A0A0L0BUH4 B4J304 A0A1J1IGR0 A0A1B6KH25 A0A1I8Q3C5 A0A1J1J7Q4 H9IYU3 U4UE05 N6TSI2 A0A0P5RNZ5 E9FST5 A0A0P5XNV6 A0A0P5LU54 T1JIM6 A0A3R7M9U9
Pubmed
26354079
22118469
26227816
24845553
25401762
20798317
+ More
21719571 21347285 21282665 20075255 28004739 24508170 30249741 20566863 25474469 17510324 26334808 27129103 26483478 20966253 12364791 14747013 17210077 20920257 23761445 25244985 18362917 19820115 24495485 25348373 25830018 17994087 7643388 10731132 12537572 12537569 15385157 25315136 15632085 22936249 26108605 19121390 23537049 21292972
21719571 21347285 21282665 20075255 28004739 24508170 30249741 20566863 25474469 17510324 26334808 27129103 26483478 20966253 12364791 14747013 17210077 20920257 23761445 25244985 18362917 19820115 24495485 25348373 25830018 17994087 7643388 10731132 12537572 12537569 15385157 25315136 15632085 22936249 26108605 19121390 23537049 21292972
EMBL
GDQN01011237
GDQN01002207
JAT79817.1
JAT88847.1
KQ459220
KPJ02873.1
+ More
ODYU01005387 SOQ46176.1 KQ459984 KPJ18993.1 AGBW02003014 OWR55588.1 JTDY01006954 KOB65570.1 GEDC01003119 JAS34179.1 NEVH01000250 PNF43871.1 KK852808 KDR16018.1 GBHO01009017 JAG34587.1 GBYB01006466 JAG76233.1 KQ982729 KYQ51563.1 KQ981374 KYN42394.1 GL453868 EFN75387.1 GL888761 EGI58109.1 KQ978231 KYM96178.1 KQ980775 KYN13235.1 LBMM01000540 KMQ98104.1 GL440609 EFN65681.1 KQ976560 KYM80593.1 ADTU01020778 GL767845 EFZ12994.1 KZ288422 PBC25956.1 KQ760129 OAD61839.1 GEZM01098871 JAV53696.1 KK107499 QOIP01000007 EZA49751.1 RLU20495.1 KQ434948 KZC12490.1 DS235875 EEB19764.1 GBBI01002466 JAC16246.1 LHQN01029188 KQ414657 KOC58745.1 KOC65739.1 GECL01002743 JAP03381.1 LJIG01001249 KRT85691.1 CH477602 EAT38438.1 GBGD01000210 JAC88679.1 GFDL01002893 JAV32152.1 GDKW01001371 JAI55224.1 JXUM01115630 KQ565901 KXJ70552.1 APCN01004639 AAAB01008948 EAA10373.3 AXCM01003267 GGFK01005731 MBW39052.1 ADMH02002171 ETN58075.1 AXCN02000979 UFQT01000031 SSX18267.1 DS497673 EFA13274.1 GAMC01017512 JAB89043.1 GDHF01026771 JAI25543.1 GAKP01017974 JAC40978.1 GBXI01003720 JAD10572.1 CH963847 EDW72639.1 CP012525 ALC44257.1 CH902618 EDV41344.1 CH933809 EDW19696.1 CCAG010018197 JXJN01023210 X80028 AE014296 AY118579 CH954178 EDV52754.1 KA646783 AFP61412.1 OUUW01000002 SPP76349.1 CH379070 EAL30250.1 CH479208 EDW31145.1 CM000363 CM002912 EDX11484.1 KMZ01168.1 CH940647 EDW70297.1 CH480826 EDW44461.1 JRES01001310 KNC23681.1 CH916366 EDV97174.1 CVRI01000050 CRK99437.1 GEBQ01029258 JAT10719.1 CVRI01000072 CRL07502.1 BABH01010805 BABH01010806 KB632314 ERL92204.1 APGK01053803 KB741235 ENN72215.1 GDIQ01107025 JAL44701.1 GL732524 EFX89764.1 GDIP01081728 LRGB01000890 JAM21987.1 KZS15328.1 GDIQ01165551 JAK86174.1 JH432213 QCYY01002360 ROT70995.1
ODYU01005387 SOQ46176.1 KQ459984 KPJ18993.1 AGBW02003014 OWR55588.1 JTDY01006954 KOB65570.1 GEDC01003119 JAS34179.1 NEVH01000250 PNF43871.1 KK852808 KDR16018.1 GBHO01009017 JAG34587.1 GBYB01006466 JAG76233.1 KQ982729 KYQ51563.1 KQ981374 KYN42394.1 GL453868 EFN75387.1 GL888761 EGI58109.1 KQ978231 KYM96178.1 KQ980775 KYN13235.1 LBMM01000540 KMQ98104.1 GL440609 EFN65681.1 KQ976560 KYM80593.1 ADTU01020778 GL767845 EFZ12994.1 KZ288422 PBC25956.1 KQ760129 OAD61839.1 GEZM01098871 JAV53696.1 KK107499 QOIP01000007 EZA49751.1 RLU20495.1 KQ434948 KZC12490.1 DS235875 EEB19764.1 GBBI01002466 JAC16246.1 LHQN01029188 KQ414657 KOC58745.1 KOC65739.1 GECL01002743 JAP03381.1 LJIG01001249 KRT85691.1 CH477602 EAT38438.1 GBGD01000210 JAC88679.1 GFDL01002893 JAV32152.1 GDKW01001371 JAI55224.1 JXUM01115630 KQ565901 KXJ70552.1 APCN01004639 AAAB01008948 EAA10373.3 AXCM01003267 GGFK01005731 MBW39052.1 ADMH02002171 ETN58075.1 AXCN02000979 UFQT01000031 SSX18267.1 DS497673 EFA13274.1 GAMC01017512 JAB89043.1 GDHF01026771 JAI25543.1 GAKP01017974 JAC40978.1 GBXI01003720 JAD10572.1 CH963847 EDW72639.1 CP012525 ALC44257.1 CH902618 EDV41344.1 CH933809 EDW19696.1 CCAG010018197 JXJN01023210 X80028 AE014296 AY118579 CH954178 EDV52754.1 KA646783 AFP61412.1 OUUW01000002 SPP76349.1 CH379070 EAL30250.1 CH479208 EDW31145.1 CM000363 CM002912 EDX11484.1 KMZ01168.1 CH940647 EDW70297.1 CH480826 EDW44461.1 JRES01001310 KNC23681.1 CH916366 EDV97174.1 CVRI01000050 CRK99437.1 GEBQ01029258 JAT10719.1 CVRI01000072 CRL07502.1 BABH01010805 BABH01010806 KB632314 ERL92204.1 APGK01053803 KB741235 ENN72215.1 GDIQ01107025 JAL44701.1 GL732524 EFX89764.1 GDIP01081728 LRGB01000890 JAM21987.1 KZS15328.1 GDIQ01165551 JAK86174.1 JH432213 QCYY01002360 ROT70995.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
UP000027135
+ More
UP000075809 UP000078541 UP000008237 UP000007755 UP000078542 UP000078492 UP000036403 UP000000311 UP000078540 UP000005205 UP000242457 UP000002358 UP000053097 UP000279307 UP000005203 UP000192223 UP000076502 UP000009046 UP000053825 UP000008820 UP000069940 UP000249989 UP000075840 UP000075902 UP000075882 UP000007062 UP000075903 UP000075883 UP000075901 UP000076407 UP000000673 UP000075886 UP000076408 UP000075920 UP000075880 UP000075881 UP000075884 UP000075885 UP000007266 UP000075900 UP000007798 UP000092553 UP000007801 UP000091820 UP000009192 UP000092444 UP000092443 UP000092460 UP000092445 UP000078200 UP000000803 UP000095301 UP000008711 UP000268350 UP000001819 UP000008744 UP000192221 UP000000304 UP000008792 UP000001292 UP000037069 UP000001070 UP000183832 UP000095300 UP000005204 UP000030742 UP000019118 UP000000305 UP000076858 UP000283509
UP000075809 UP000078541 UP000008237 UP000007755 UP000078542 UP000078492 UP000036403 UP000000311 UP000078540 UP000005205 UP000242457 UP000002358 UP000053097 UP000279307 UP000005203 UP000192223 UP000076502 UP000009046 UP000053825 UP000008820 UP000069940 UP000249989 UP000075840 UP000075902 UP000075882 UP000007062 UP000075903 UP000075883 UP000075901 UP000076407 UP000000673 UP000075886 UP000076408 UP000075920 UP000075880 UP000075881 UP000075884 UP000075885 UP000007266 UP000075900 UP000007798 UP000092553 UP000007801 UP000091820 UP000009192 UP000092444 UP000092443 UP000092460 UP000092445 UP000078200 UP000000803 UP000095301 UP000008711 UP000268350 UP000001819 UP000008744 UP000192221 UP000000304 UP000008792 UP000001292 UP000037069 UP000001070 UP000183832 UP000095300 UP000005204 UP000030742 UP000019118 UP000000305 UP000076858 UP000283509
Interpro
Gene 3D
ProteinModelPortal
A0A1E1VYL1
A0A194QBF4
A0A2H1VZQ4
A0A194RME1
A0A212FPB6
A0A0L7KR32
+ More
A0A1B6E8D2 A0A2J7RSR7 A0A067R1Z2 A0A0A9YNG5 A0A0C9RHN7 A0A151WUN7 A0A151JZS4 E2C9I5 F4X615 A0A195C7Q6 A0A151IY57 A0A0J7L6I5 E2ALH9 A0A151I1Z4 A0A158NMS9 E9J2G9 A0A2A3E3X8 K7IXQ7 A0A310SQH9 A0A1Y1K2U0 A0A026W2C6 A0A088AFJ6 A0A1W4WP41 A0A154PMQ3 E0W2A8 A0A023F447 A0A0L7QJK1 A0A0V0G7U7 A0A0T6BES9 Q16V64 A0A069DZQ9 A0A1Q3FXH3 A0A0N7Z970 A0A182H761 A0A2C9GQZ7 A0A182TK65 A0A182KNQ8 Q7PPI1 A0A182VI02 A0A182M787 A0A2M4AEB2 A0A182SLJ9 A0A182X2H7 W5J5D3 A0A182QSA6 A0A336LPG5 A0A182YKR9 A0A182WM09 A0A182JBE8 A0A182K4N4 A0A182N1Z7 A0A182P5G5 D7EI65 A0A182RAL7 W8AWC4 A0A0K8UG50 A0A034VHD8 A0A0A1XHY9 B4MND0 A0A0M4F0H1 B3MB43 A0A1A9WSP3 B4KX53 A0A1B0FKC3 A0A1A9XGF2 A0A1B0BZM6 A0A1A9ZKP0 A0A1A9UFU4 P55162 A0A1I8M685 B3NJ37 T1PFW3 A0A3B0JRM4 Q29E13 B4H430 A0A1W4UZ08 B4QKR9 B4LFH9 B4IAZ8 A0A0L0BUH4 B4J304 A0A1J1IGR0 A0A1B6KH25 A0A1I8Q3C5 A0A1J1J7Q4 H9IYU3 U4UE05 N6TSI2 A0A0P5RNZ5 E9FST5 A0A0P5XNV6 A0A0P5LU54 T1JIM6 A0A3R7M9U9
A0A1B6E8D2 A0A2J7RSR7 A0A067R1Z2 A0A0A9YNG5 A0A0C9RHN7 A0A151WUN7 A0A151JZS4 E2C9I5 F4X615 A0A195C7Q6 A0A151IY57 A0A0J7L6I5 E2ALH9 A0A151I1Z4 A0A158NMS9 E9J2G9 A0A2A3E3X8 K7IXQ7 A0A310SQH9 A0A1Y1K2U0 A0A026W2C6 A0A088AFJ6 A0A1W4WP41 A0A154PMQ3 E0W2A8 A0A023F447 A0A0L7QJK1 A0A0V0G7U7 A0A0T6BES9 Q16V64 A0A069DZQ9 A0A1Q3FXH3 A0A0N7Z970 A0A182H761 A0A2C9GQZ7 A0A182TK65 A0A182KNQ8 Q7PPI1 A0A182VI02 A0A182M787 A0A2M4AEB2 A0A182SLJ9 A0A182X2H7 W5J5D3 A0A182QSA6 A0A336LPG5 A0A182YKR9 A0A182WM09 A0A182JBE8 A0A182K4N4 A0A182N1Z7 A0A182P5G5 D7EI65 A0A182RAL7 W8AWC4 A0A0K8UG50 A0A034VHD8 A0A0A1XHY9 B4MND0 A0A0M4F0H1 B3MB43 A0A1A9WSP3 B4KX53 A0A1B0FKC3 A0A1A9XGF2 A0A1B0BZM6 A0A1A9ZKP0 A0A1A9UFU4 P55162 A0A1I8M685 B3NJ37 T1PFW3 A0A3B0JRM4 Q29E13 B4H430 A0A1W4UZ08 B4QKR9 B4LFH9 B4IAZ8 A0A0L0BUH4 B4J304 A0A1J1IGR0 A0A1B6KH25 A0A1I8Q3C5 A0A1J1J7Q4 H9IYU3 U4UE05 N6TSI2 A0A0P5RNZ5 E9FST5 A0A0P5XNV6 A0A0P5LU54 T1JIM6 A0A3R7M9U9
PDB
4N78
E-value=0,
Score=3659
Ontologies
GO
PANTHER
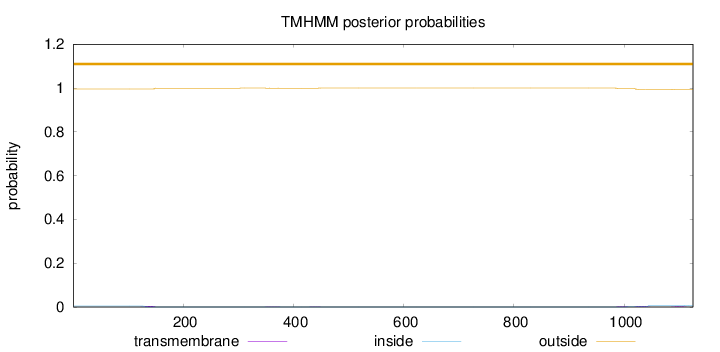
Topology
Subcellular location
Cell membrane
Length:
1125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27054
Exp number, first 60 AAs:
0.00088
Total prob of N-in:
0.00457
outside
1 - 1125
Population Genetic Test Statistics
Pi
224.174433
Theta
211.458761
Tajima's D
0.223451
CLR
0.145437
CSRT
0.428828558572071
Interpretation
Uncertain
