Gene
KWMTBOMO05230
Pre Gene Modal
BGIBMGA002480
Annotation
PREDICTED:_disintegrin_and_metalloproteinase_domain-containing_protein_10_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.337
Sequence
CDS
ATGTTTAGTGAAATTGGATACATTACATTTCTTCTACTTGCTTTACCATACATTGAAAGTGCATCTCGCGTTCGTCTTAACGAATACATCGAGCACTTCGAAACGCTAGACTACGATCCAGATGATTTGCATCATCAACACTTGCGAGCTCGACGCTCCACGGATTCTCAACGAGAACTGCGACTCGACTTCAAAGCGCACGACAGGAAATTCAAGTTGCGTTTGCGACGAGACTTGTCCGCTTTCAGCGATGACTTTAAGGTGGAAGGCTCACAAGGACAAACACACGAAGTTGACTCATCGCACATTTACAGCGGAAAACTGGCCGGCGACCCAGACTCGACTGTTTTCGGATCGGTGACGGAAGGTGTCTTCGAGGGGAAGATAATCGCCAAAGACGGTTCTTACTACGTGGAACACGCAAGAAGATACTTCCCACCTAACAACACGAAGACACGAGGCCACTCCGTTATTTACAAAGATAGTGATGTCATTGATCCGTTTGCTAACAAAAGGCACGGTCACGTCGGCGGATGCGGTATAACCGATGAAGTGGTCCAATGGATGGAACGCATTCAGAACTCTGGAGTTGACGATGAATCTCCCACGTCGCCGGCATCTTCACAGTCCACGCCTACTGTCGCAACTCCACCACACACTCACACCTCCGGCTCGCCCTCGCCTCATCCCAATAGCTTAAGAGATGATATGCCAAGACATTGGGATTCACATTTTCACAATAAATATTCGCGGTCCGCGAACACGGGCGAGCATGCGCGTTCGCGTCGAGCAGCGCCGTATGACAACCGCAACACGTGCTCGCTGTTCATACAAACTGATCCCTTAATATGGAAGCATATTCGTGAGGGATTTCCCGACCATAATGTGCCAAGTAAAAAGAACGAAGTCGACACGAAGACTCGGGAAGAGATCCTGTCTCTCATATCGCATCACGTGACTGCCGTCAACTACATCTACGGCAGGGCCAAGTTCGACGGCAGATTCGACCACAGGGACATCAAGTTTGAAGTGCAGAGAATAAAGATCGACGACAACTCACTGTGTAACAATCGTTATCCGAACGAAATGAATCAGTTCTGTTTAACGAACGTTGACGTTTCGAATTTCCTGAATCTCCACTCGCTTGGCAACCACGAGGACTTCTGTCTGGCCTACGTGTTCACGTACAGAGACTTCACCGGGGGGACCCTAGGTCTGGCATGGGTGGCGAGCGCGAGCGGCGCGTCGGGCGGCATCTGCGAGAAGTACAAGACGTACACGGAGACGATCGGCGGCATGTACCAGAGCACCAAGCGCTCGCTCAACACGGGCATCATCACGTTCGTCAACTACAACAGCAGGGTGCCGCCCAAGGTCAGTCAGCTCACGCTGGCGCACGAGATCGGGCACAACTTCGGCTCGCCGCACGACTACCCCTCGGAGTGCCGGCCGGGGGGCCAGCAGGGCAACTACATAATGTTCGCGTCGGCGACGTCCGGCGACCGGCCCAACAACAGCCGGTTCTCGTCGTGCAGCACCGGCAACATCAGCGCCGTGCTGGACGCCGTCAGGGACGGGCGCAAGCGGAACTGCCTCACCGCCAGCGCTGGCGCCTTCTGCGGGAACAAGATCGTCGAGCACGGCGAGGAGTGCGACTGCGGCTACGACGACATCGAGTGCACGGACCGGTGCTGCTACCCGCGCCAGGTCGGCGCCTACGACCGCGAGAGGAACGTCACCGCCAAGGGCTGCACCAGACGCGCCGACACAGAGTGCAGTCCCTCCCAGGGTCCGTGCTGCAACGCGCAGACGTGCCGGTTCGTGGTGGCGGCGCGCAACCAGACGTGTCGCGAGGCGGCCGAGTGCAGCCACGCGGCCGTCTGCTCCGGGGACTCCGCCGATTGCCCCGAGCCCAGTGCCATGCCCAATCACACCAAATGTAACAACGGCACGCAACTGTGCAAGGACGGCGAGTGCAGCGAGTCGATATGTCGCGCGTGGAACATGTCGGTGTGCTACCTGTCGGCCAGCGCGGCCCGCGGCGCCGGCGTCACGGCGCAGCCCAACCGCCGCGCGCTGTGCCAGCTCGCGTGCCAGCAGGGCTCCGACCCCGACTCGTGCCGCAGCACCGCGGACTTCGCCAGCGAGGTCGGCCTGCCCACCGGCGGGATCAGCCTCCTGCCCGGCTCGCCCTGCGACAACTTCCAGGGCTACTGCGACGTGTTCCTGAAGTGCCGCGCCGTGGACGCTGAGGGTCCGCTGGTGCGGCTTAAGAACCTCCTGCTGAACCGCGCCACGCTGCAGTCGGTGCAGGCCTGGGTCACGGAGATGTGGTGGGCGGTGCTGCTGGCCGGGGTGGCGCTCGTCGTGTGCATGGGAGCATTCGTCAAGTGCTGTGCCGTGCATACGCCCTCATCGAATCCGAAACGACCGCCCGCCAGACGGCTCTCGGAGACGCTGCGCGGCCCCATGAACACGCTACGGCGCATGCGCGGACCACGCGGGACTCCGGCTCGGCACCCGCAACGCCGCGCCCGCACGCACAAGGGCTACGCGCCGCCACACACGTACGCGCATCGAGACCTGCCGACGGCGCCGGCTGGACCCAGCGGGCGGCGGGGCGAGCCGTACGCCAACGCGTACCCCGCGGCGTACGAGATGCGGCCCGTGCACAAGATATCGATCCATGTTCCGCTGAGCTTAGCTGCTGACGTGCGACACACAAAACACGGACGGACACTTGAAATTATTTTTATACGATGA
Protein
MFSEIGYITFLLLALPYIESASRVRLNEYIEHFETLDYDPDDLHHQHLRARRSTDSQRELRLDFKAHDRKFKLRLRRDLSAFSDDFKVEGSQGQTHEVDSSHIYSGKLAGDPDSTVFGSVTEGVFEGKIIAKDGSYYVEHARRYFPPNNTKTRGHSVIYKDSDVIDPFANKRHGHVGGCGITDEVVQWMERIQNSGVDDESPTSPASSQSTPTVATPPHTHTSGSPSPHPNSLRDDMPRHWDSHFHNKYSRSANTGEHARSRRAAPYDNRNTCSLFIQTDPLIWKHIREGFPDHNVPSKKNEVDTKTREEILSLISHHVTAVNYIYGRAKFDGRFDHRDIKFEVQRIKIDDNSLCNNRYPNEMNQFCLTNVDVSNFLNLHSLGNHEDFCLAYVFTYRDFTGGTLGLAWVASASGASGGICEKYKTYTETIGGMYQSTKRSLNTGIITFVNYNSRVPPKVSQLTLAHEIGHNFGSPHDYPSECRPGGQQGNYIMFASATSGDRPNNSRFSSCSTGNISAVLDAVRDGRKRNCLTASAGAFCGNKIVEHGEECDCGYDDIECTDRCCYPRQVGAYDRERNVTAKGCTRRADTECSPSQGPCCNAQTCRFVVAARNQTCREAAECSHAAVCSGDSADCPEPSAMPNHTKCNNGTQLCKDGECSESICRAWNMSVCYLSASAARGAGVTAQPNRRALCQLACQQGSDPDSCRSTADFASEVGLPTGGISLLPGSPCDNFQGYCDVFLKCRAVDAEGPLVRLKNLLLNRATLQSVQAWVTEMWWAVLLAGVALVVCMGAFVKCCAVHTPSSNPKRPPARRLSETLRGPMNTLRRMRGPRGTPARHPQRRARTHKGYAPPHTYAHRDLPTAPAGPSGRRGEPYANAYPAAYEMRPVHKISIHVPLSLAADVRHTKHGRTLEIIFIR
Summary
Uniprot
H9IYZ5
A0A1Y1LVQ2
A0A1Y1LWV9
A0A194QBN5
A0A1W4WQ48
A0A1W4WQD5
+ More
A0A1W4WE84 D7EI66 A0A2P8Z1A2 A0A1L2JAW4 A0A1B6DQY0 A0A194RNA2 A0A224XJQ1 A0A088AEU3 A0A1B6G5D5 A0A310SGU7 A0A0T6AZV6 A0A336M645 A0A336M3J2 A0A1L8E3C5 A0A1L8E3E1 A0A0M8ZUE1 U4UII6 A0A1B6KFK6 A0A1B6LPU7 A0A348G5Z3 A0A212FPG3 E2C9I4 E2ALI0 A0A2A3E396 A0A067RM49 A0A336LXA8 A0A0C9R6L6 A0A0J7L6D0 A0A336LPH5 A0A195C5Q6 A0A0C9RB19 A0A158NMT0 A0A151IY61 A0A3L8DJ82 F4X616 A0A151WUP5 A0A0A9WUF9 A0A2R7VUZ5 E0VKA6 A0A2J7RAJ7 A0A1J1I5P2 K7IXP2 A0A2S2QT51 A0A182Y8H6 A0A182LY44 A0A182VTN7 A0A2H8TK40 A0A2S2N6P3 X1WN16 A0A0P5BLC8 A0A0P5FZF1 E9FVI8 A0A0P5G280 A0A0P5M688 A0A0N7ZNW6 A0A0N8BMQ7 A0A0P5J517 A0A0P5LT59 A0A0P5L8H4 A0A0P5IDH4 A0A0P5E8I2 A0A0P4ZR40 A0A0N7ZYA3 A0A0P5BED2 A0A0P4YP75 A0A0P4YB00 A0A0P4ZGM3 A0A0P5I3Z3 A0A0P5KUE6 A0A0P5GN01 A0A0P5ILN0 A0A0P5D675 A0A0P5BJK6 A0A0P4XKN9 A0A0P5KMB7 A0A0P5BBU2 A0A0P5JG26 A0A0P5BLI7 A0A0P4WS71 A0A0P4XQ18 A0A0P6GMY2 A0A0P5D0X4 A0A0P5AAF3 A0A0N8EM46 A0A0P4YJC1 A0A0P5TV19 A0A0P4XTW3 A0A0P5JXN0 A0A0P5C6K4 A0A0N7ZN60 A0A0P5L4G9 A0A0P5HU83 A0A1B6JDR8
A0A1W4WE84 D7EI66 A0A2P8Z1A2 A0A1L2JAW4 A0A1B6DQY0 A0A194RNA2 A0A224XJQ1 A0A088AEU3 A0A1B6G5D5 A0A310SGU7 A0A0T6AZV6 A0A336M645 A0A336M3J2 A0A1L8E3C5 A0A1L8E3E1 A0A0M8ZUE1 U4UII6 A0A1B6KFK6 A0A1B6LPU7 A0A348G5Z3 A0A212FPG3 E2C9I4 E2ALI0 A0A2A3E396 A0A067RM49 A0A336LXA8 A0A0C9R6L6 A0A0J7L6D0 A0A336LPH5 A0A195C5Q6 A0A0C9RB19 A0A158NMT0 A0A151IY61 A0A3L8DJ82 F4X616 A0A151WUP5 A0A0A9WUF9 A0A2R7VUZ5 E0VKA6 A0A2J7RAJ7 A0A1J1I5P2 K7IXP2 A0A2S2QT51 A0A182Y8H6 A0A182LY44 A0A182VTN7 A0A2H8TK40 A0A2S2N6P3 X1WN16 A0A0P5BLC8 A0A0P5FZF1 E9FVI8 A0A0P5G280 A0A0P5M688 A0A0N7ZNW6 A0A0N8BMQ7 A0A0P5J517 A0A0P5LT59 A0A0P5L8H4 A0A0P5IDH4 A0A0P5E8I2 A0A0P4ZR40 A0A0N7ZYA3 A0A0P5BED2 A0A0P4YP75 A0A0P4YB00 A0A0P4ZGM3 A0A0P5I3Z3 A0A0P5KUE6 A0A0P5GN01 A0A0P5ILN0 A0A0P5D675 A0A0P5BJK6 A0A0P4XKN9 A0A0P5KMB7 A0A0P5BBU2 A0A0P5JG26 A0A0P5BLI7 A0A0P4WS71 A0A0P4XQ18 A0A0P6GMY2 A0A0P5D0X4 A0A0P5AAF3 A0A0N8EM46 A0A0P4YJC1 A0A0P5TV19 A0A0P4XTW3 A0A0P5JXN0 A0A0P5C6K4 A0A0N7ZN60 A0A0P5L4G9 A0A0P5HU83 A0A1B6JDR8
Pubmed
EMBL
BABH01010800
BABH01010801
BABH01010802
BABH01010803
BABH01010804
BABH01010805
+ More
GEZM01049082 JAV76115.1 GEZM01049081 JAV76116.1 KQ459220 KPJ02872.1 KQ971379 EFA13242.2 PYGN01000245 PSN50272.1 KX388389 AOT22059.1 GEDC01012649 GEDC01009210 JAS24649.1 JAS28088.1 KQ459984 KPJ18992.1 GFTR01008187 JAW08239.1 GECZ01026790 GECZ01012132 JAS42979.1 JAS57637.1 KQ760129 OAD61840.1 LJIG01016393 KRT80735.1 UFQT01000502 SSX24821.1 SSX24822.1 GFDF01000844 JAV13240.1 GFDF01000843 JAV13241.1 KQ435882 KOX69803.1 KB632314 ERL92203.1 GEBQ01029764 JAT10213.1 GEBQ01014217 JAT25760.1 FX985531 BBF97866.1 AGBW02003014 OWR55590.1 GL453868 EFN75386.1 GL440609 EFN65682.1 KZ288422 PBC25954.1 KK852427 KDR24098.1 UFQS01000031 UFQT01000031 SSW97892.1 SSX18278.1 GBYB01011974 JAG81741.1 LBMM01000540 KMQ98103.1 SSW97891.1 SSX18277.1 KQ978231 KYM96177.1 GBYB01010307 JAG80074.1 ADTU01020779 KQ980775 KYN13236.1 QOIP01000007 RLU20494.1 GL888761 EGI58110.1 KQ982729 KYQ51564.1 GBHO01035104 GBHO01035103 GBHO01035102 GBRD01004395 GBRD01004394 JAG08500.1 JAG08501.1 JAG08502.1 JAG61426.1 KK854102 PTY11324.1 DS235241 EEB13812.1 NEVH01006567 PNF37858.1 CVRI01000038 CRK94254.1 GGMS01011497 MBY80700.1 AXCM01001785 GFXV01002604 MBW14409.1 GGMR01000252 MBY12871.1 ABLF02028257 GDIP01182909 JAJ40493.1 GDIQ01253537 JAJ98187.1 GL732525 EFX89096.1 GDIQ01252239 JAJ99485.1 GDIQ01184098 JAK67627.1 GDIP01227009 JAI96392.1 GDIQ01162603 JAK89122.1 GDIQ01203090 JAK48635.1 GDIQ01190206 JAK61519.1 GDIQ01178602 JAK73123.1 GDIQ01215983 JAK35742.1 GDIP01163461 JAJ59941.1 GDIP01209858 JAJ13544.1 GDIP01200713 JAJ22689.1 GDIP01185560 JAJ37842.1 GDIP01224556 JAI98845.1 GDIP01231156 JAI92245.1 GDIP01212862 JAJ10540.1 GDIQ01220640 JAK31085.1 GDIQ01227036 GDIQ01204889 JAK46836.1 GDIQ01244943 JAK06782.1 GDIQ01212381 JAK39344.1 GDIP01162411 JAJ60991.1 GDIP01184111 JAJ39291.1 GDIP01240511 JAI82890.1 GDIQ01183252 JAK68473.1 GDIP01186667 JAJ36735.1 GDIQ01198994 JAK52731.1 GDIP01197811 JAJ25591.1 GDIP01251503 GDIP01112567 JAI71898.1 GDIP01238014 GDIP01227008 JAI85387.1 GDIQ01041558 JAN53179.1 GDIP01164410 JAJ58992.1 GDIP01201909 JAJ21493.1 GDIQ01013300 JAN81437.1 GDIP01229056 JAI94345.1 GDIP01124066 JAL79648.1 GDIP01236662 JAI86739.1 GDIQ01197594 JAK54131.1 GDIP01179058 JAJ44344.1 GDIP01229057 JAI94344.1 GDIQ01175098 JAK76627.1 GDIQ01228025 JAK23700.1 GECU01010285 JAS97421.1
GEZM01049082 JAV76115.1 GEZM01049081 JAV76116.1 KQ459220 KPJ02872.1 KQ971379 EFA13242.2 PYGN01000245 PSN50272.1 KX388389 AOT22059.1 GEDC01012649 GEDC01009210 JAS24649.1 JAS28088.1 KQ459984 KPJ18992.1 GFTR01008187 JAW08239.1 GECZ01026790 GECZ01012132 JAS42979.1 JAS57637.1 KQ760129 OAD61840.1 LJIG01016393 KRT80735.1 UFQT01000502 SSX24821.1 SSX24822.1 GFDF01000844 JAV13240.1 GFDF01000843 JAV13241.1 KQ435882 KOX69803.1 KB632314 ERL92203.1 GEBQ01029764 JAT10213.1 GEBQ01014217 JAT25760.1 FX985531 BBF97866.1 AGBW02003014 OWR55590.1 GL453868 EFN75386.1 GL440609 EFN65682.1 KZ288422 PBC25954.1 KK852427 KDR24098.1 UFQS01000031 UFQT01000031 SSW97892.1 SSX18278.1 GBYB01011974 JAG81741.1 LBMM01000540 KMQ98103.1 SSW97891.1 SSX18277.1 KQ978231 KYM96177.1 GBYB01010307 JAG80074.1 ADTU01020779 KQ980775 KYN13236.1 QOIP01000007 RLU20494.1 GL888761 EGI58110.1 KQ982729 KYQ51564.1 GBHO01035104 GBHO01035103 GBHO01035102 GBRD01004395 GBRD01004394 JAG08500.1 JAG08501.1 JAG08502.1 JAG61426.1 KK854102 PTY11324.1 DS235241 EEB13812.1 NEVH01006567 PNF37858.1 CVRI01000038 CRK94254.1 GGMS01011497 MBY80700.1 AXCM01001785 GFXV01002604 MBW14409.1 GGMR01000252 MBY12871.1 ABLF02028257 GDIP01182909 JAJ40493.1 GDIQ01253537 JAJ98187.1 GL732525 EFX89096.1 GDIQ01252239 JAJ99485.1 GDIQ01184098 JAK67627.1 GDIP01227009 JAI96392.1 GDIQ01162603 JAK89122.1 GDIQ01203090 JAK48635.1 GDIQ01190206 JAK61519.1 GDIQ01178602 JAK73123.1 GDIQ01215983 JAK35742.1 GDIP01163461 JAJ59941.1 GDIP01209858 JAJ13544.1 GDIP01200713 JAJ22689.1 GDIP01185560 JAJ37842.1 GDIP01224556 JAI98845.1 GDIP01231156 JAI92245.1 GDIP01212862 JAJ10540.1 GDIQ01220640 JAK31085.1 GDIQ01227036 GDIQ01204889 JAK46836.1 GDIQ01244943 JAK06782.1 GDIQ01212381 JAK39344.1 GDIP01162411 JAJ60991.1 GDIP01184111 JAJ39291.1 GDIP01240511 JAI82890.1 GDIQ01183252 JAK68473.1 GDIP01186667 JAJ36735.1 GDIQ01198994 JAK52731.1 GDIP01197811 JAJ25591.1 GDIP01251503 GDIP01112567 JAI71898.1 GDIP01238014 GDIP01227008 JAI85387.1 GDIQ01041558 JAN53179.1 GDIP01164410 JAJ58992.1 GDIP01201909 JAJ21493.1 GDIQ01013300 JAN81437.1 GDIP01229056 JAI94345.1 GDIP01124066 JAL79648.1 GDIP01236662 JAI86739.1 GDIQ01197594 JAK54131.1 GDIP01179058 JAJ44344.1 GDIP01229057 JAI94344.1 GDIQ01175098 JAK76627.1 GDIQ01228025 JAK23700.1 GECU01010285 JAS97421.1
Proteomes
UP000005204
UP000053268
UP000192223
UP000007266
UP000245037
UP000053240
+ More
UP000005203 UP000053105 UP000030742 UP000007151 UP000008237 UP000000311 UP000242457 UP000027135 UP000036403 UP000078542 UP000005205 UP000078492 UP000279307 UP000007755 UP000075809 UP000009046 UP000235965 UP000183832 UP000002358 UP000076408 UP000075883 UP000075920 UP000007819 UP000000305
UP000005203 UP000053105 UP000030742 UP000007151 UP000008237 UP000000311 UP000242457 UP000027135 UP000036403 UP000078542 UP000005205 UP000078492 UP000279307 UP000007755 UP000075809 UP000009046 UP000235965 UP000183832 UP000002358 UP000076408 UP000075883 UP000075920 UP000007819 UP000000305
PRIDE
Interpro
SUPFAM
SSF57552
SSF57552
Gene 3D
ProteinModelPortal
H9IYZ5
A0A1Y1LVQ2
A0A1Y1LWV9
A0A194QBN5
A0A1W4WQ48
A0A1W4WQD5
+ More
A0A1W4WE84 D7EI66 A0A2P8Z1A2 A0A1L2JAW4 A0A1B6DQY0 A0A194RNA2 A0A224XJQ1 A0A088AEU3 A0A1B6G5D5 A0A310SGU7 A0A0T6AZV6 A0A336M645 A0A336M3J2 A0A1L8E3C5 A0A1L8E3E1 A0A0M8ZUE1 U4UII6 A0A1B6KFK6 A0A1B6LPU7 A0A348G5Z3 A0A212FPG3 E2C9I4 E2ALI0 A0A2A3E396 A0A067RM49 A0A336LXA8 A0A0C9R6L6 A0A0J7L6D0 A0A336LPH5 A0A195C5Q6 A0A0C9RB19 A0A158NMT0 A0A151IY61 A0A3L8DJ82 F4X616 A0A151WUP5 A0A0A9WUF9 A0A2R7VUZ5 E0VKA6 A0A2J7RAJ7 A0A1J1I5P2 K7IXP2 A0A2S2QT51 A0A182Y8H6 A0A182LY44 A0A182VTN7 A0A2H8TK40 A0A2S2N6P3 X1WN16 A0A0P5BLC8 A0A0P5FZF1 E9FVI8 A0A0P5G280 A0A0P5M688 A0A0N7ZNW6 A0A0N8BMQ7 A0A0P5J517 A0A0P5LT59 A0A0P5L8H4 A0A0P5IDH4 A0A0P5E8I2 A0A0P4ZR40 A0A0N7ZYA3 A0A0P5BED2 A0A0P4YP75 A0A0P4YB00 A0A0P4ZGM3 A0A0P5I3Z3 A0A0P5KUE6 A0A0P5GN01 A0A0P5ILN0 A0A0P5D675 A0A0P5BJK6 A0A0P4XKN9 A0A0P5KMB7 A0A0P5BBU2 A0A0P5JG26 A0A0P5BLI7 A0A0P4WS71 A0A0P4XQ18 A0A0P6GMY2 A0A0P5D0X4 A0A0P5AAF3 A0A0N8EM46 A0A0P4YJC1 A0A0P5TV19 A0A0P4XTW3 A0A0P5JXN0 A0A0P5C6K4 A0A0N7ZN60 A0A0P5L4G9 A0A0P5HU83 A0A1B6JDR8
A0A1W4WE84 D7EI66 A0A2P8Z1A2 A0A1L2JAW4 A0A1B6DQY0 A0A194RNA2 A0A224XJQ1 A0A088AEU3 A0A1B6G5D5 A0A310SGU7 A0A0T6AZV6 A0A336M645 A0A336M3J2 A0A1L8E3C5 A0A1L8E3E1 A0A0M8ZUE1 U4UII6 A0A1B6KFK6 A0A1B6LPU7 A0A348G5Z3 A0A212FPG3 E2C9I4 E2ALI0 A0A2A3E396 A0A067RM49 A0A336LXA8 A0A0C9R6L6 A0A0J7L6D0 A0A336LPH5 A0A195C5Q6 A0A0C9RB19 A0A158NMT0 A0A151IY61 A0A3L8DJ82 F4X616 A0A151WUP5 A0A0A9WUF9 A0A2R7VUZ5 E0VKA6 A0A2J7RAJ7 A0A1J1I5P2 K7IXP2 A0A2S2QT51 A0A182Y8H6 A0A182LY44 A0A182VTN7 A0A2H8TK40 A0A2S2N6P3 X1WN16 A0A0P5BLC8 A0A0P5FZF1 E9FVI8 A0A0P5G280 A0A0P5M688 A0A0N7ZNW6 A0A0N8BMQ7 A0A0P5J517 A0A0P5LT59 A0A0P5L8H4 A0A0P5IDH4 A0A0P5E8I2 A0A0P4ZR40 A0A0N7ZYA3 A0A0P5BED2 A0A0P4YP75 A0A0P4YB00 A0A0P4ZGM3 A0A0P5I3Z3 A0A0P5KUE6 A0A0P5GN01 A0A0P5ILN0 A0A0P5D675 A0A0P5BJK6 A0A0P4XKN9 A0A0P5KMB7 A0A0P5BBU2 A0A0P5JG26 A0A0P5BLI7 A0A0P4WS71 A0A0P4XQ18 A0A0P6GMY2 A0A0P5D0X4 A0A0P5AAF3 A0A0N8EM46 A0A0P4YJC1 A0A0P5TV19 A0A0P4XTW3 A0A0P5JXN0 A0A0P5C6K4 A0A0N7ZN60 A0A0P5L4G9 A0A0P5HU83 A0A1B6JDR8
PDB
6BE6
E-value=3.26257e-102,
Score=953
Ontologies
GO
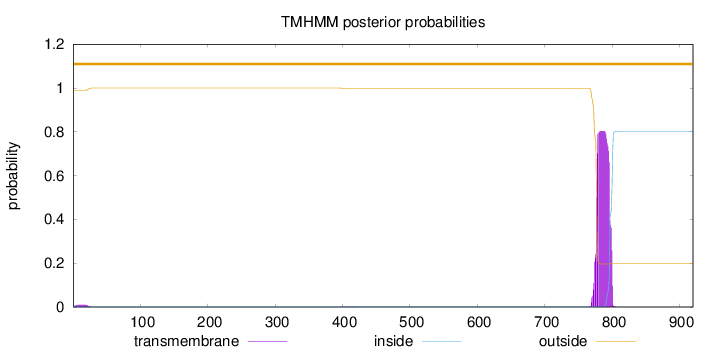
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.699245
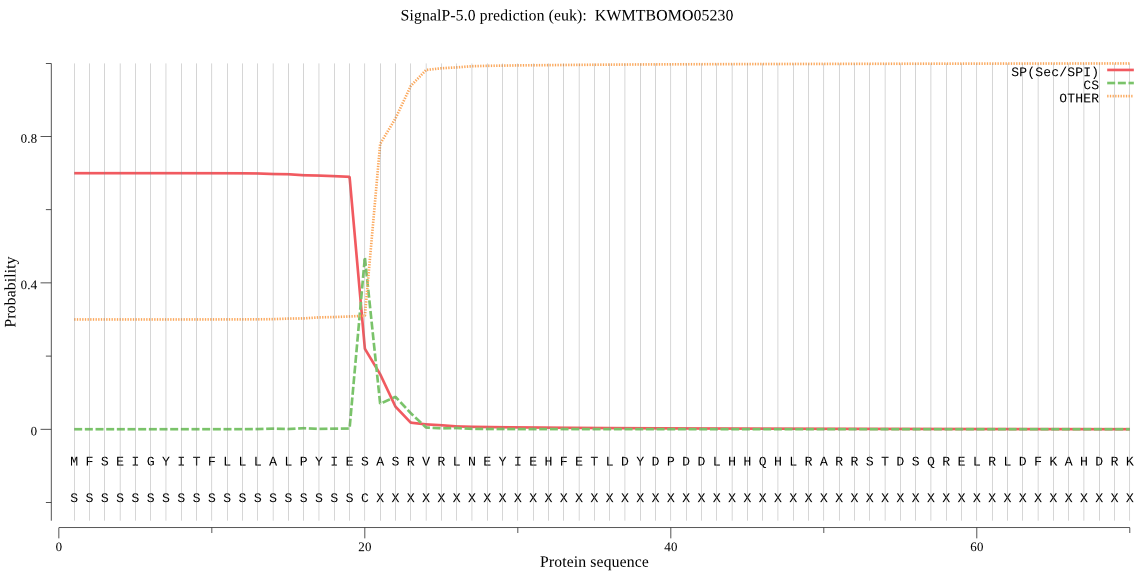
Length:
920
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.93499
Exp number, first 60 AAs:
0.19449
Total prob of N-in:
0.01030
outside
1 - 920
Population Genetic Test Statistics
Pi
223.548609
Theta
174.184129
Tajima's D
0.456657
CLR
0.796174
CSRT
0.501174941252937
Interpretation
Uncertain
