Pre Gene Modal
BGIBMGA002478
Annotation
methylthioribose-1-phosphate_isomerase_[Bombyx_mori]
Full name
Methylthioribose-1-phosphate isomerase
Alternative Name
S-methyl-5-thioribose-1-phosphate isomerase
Translation initiation factor eIF-2B subunit alpha/beta/delta-like protein
Translation initiation factor eIF-2B subunit alpha/beta/delta-like protein
Location in the cell
Cytoplasmic Reliability : 1.502 Mitochondrial Reliability : 1.306
Sequence
CDS
ATGAGTTTAGAATCCATAAAATATACTAGAGGTAGTCTAGAGATCTTAGATCAATTATTATTGCCTTTACAAACGAGATATATCAAAGTGCGAGGTGTTGAGGATGGTTGGAAAGTAATAAATAAAATGCAGGTCCGTGGAGCACCAGCCATAGCCATAGTGGGTTGCTTATCACTAGCCGTTGAATTATCACCTGACAATGAATCTTCAAAAAAAAACATGAGGCAAGAGATCGAAGGTAAACTTAACTATCTGGTGTCTGCCAGGCCAACAGCTGTAAATATAAAATTGGCAGCTGATGAGCTGATAAATTTGGCAAATACACTTTGTGCTGACGACAGTATTTCAGCTGAAATTTTTAAAGAAAGGTTTATAGGTAGCATAGAGGATATGCTCACTAAAGATATACATGATAACAAAGCAATAGGAAGTTTTGGATGTGAAGCTATATTAAAAAATATAGATGGTGATAGTCCAGTCAGAGTGCTTACACACTGTAATACAGGATCCTTAGCTACAGCCGGATATGGCACTGCTTTAGGAGTTATCAGATCATTACATGCCACTAAAAGATTAGAGCATGTCTTCTGCACTGAGACAAGGCCATATAATCAAGGAGCAAGACTGACTGCGTATGAACTAGTCCACGAGAAAATCCCTTCTACATTAATTGTAGATAGTATGGTCTCTGCTCTTATGCACACCAGGAAAATACATGCAGTCATTGTTGGTGCTGACAGGGTTGCCGCTAATGGGGATACAGCAAATAAAATTGGAACGTATCAAATAGCTATTGTAGCAAAATATCATGATGTACCCTTCTACGTAGCGGCTCCGCTCACATCCATAGACATGTCTTTACCATATGGAGAAAAAATTAAAATTGAAGAACGGCCCGATAGGGAAATGACTCATATCGGAGAACATAGAATAGCTGCACCAGGCATAAATTGCTGGAACCCTTCATTTGACGTAACACCGGCATCTCTAATAGCAGGCATCATAACTGAAAAAGGTGTATTTGCTCCTGACAATCTAAAAAGTGTTAAAACCACGTAA
Protein
MSLESIKYTRGSLEILDQLLLPLQTRYIKVRGVEDGWKVINKMQVRGAPAIAIVGCLSLAVELSPDNESSKKNMRQEIEGKLNYLVSARPTAVNIKLAADELINLANTLCADDSISAEIFKERFIGSIEDMLTKDIHDNKAIGSFGCEAILKNIDGDSPVRVLTHCNTGSLATAGYGTALGVIRSLHATKRLEHVFCTETRPYNQGARLTAYELVHEKIPSTLIVDSMVSALMHTRKIHAVIVGADRVAANGDTANKIGTYQIAIVAKYHDVPFYVAAPLTSIDMSLPYGEKIKIEERPDREMTHIGEHRIAAPGINCWNPSFDVTPASLIAGIITEKGVFAPDNLKSVKTT
Summary
Description
Catalyzes the interconversion of methylthioribose-1-phosphate (MTR-1-P) into methylthioribulose-1-phosphate (MTRu-1-P).
Catalytic Activity
S-methyl-5-thio-alpha-D-ribose 1-phosphate = 5-methylsulfanyl-D-ribulose 1-phosphate
Similarity
Belongs to the eIF-2B alpha/beta/delta subunits family. MtnA subfamily.
Belongs to the eIF-2B alpha/beta/delta subunits family.
Belongs to the eIF-2B alpha/beta/delta subunits family.
Keywords
Amino-acid biosynthesis
Complete proteome
Cytoplasm
Isomerase
Methionine biosynthesis
Nucleus
Reference proteome
Feature
chain Methylthioribose-1-phosphate isomerase
Uniprot
H9IYZ3
Q0ZB81
A0A194RNT7
A0A2H1V342
A0A212FPF1
A0A194QHF3
+ More
A0A0L0CRS8 A0A336M6Q7 D3TPX8 A0A1A9V7L8 A0A1A9YPY7 A0A1I8MNW6 A0A1I8P818 A0A0K8TZJ1 A0A0A1XPU2 W8ATZ5 B4NG41 A0A3B0JLB6 A0A3S2L2C5 A0A0P9C7L9 B3M098 A0A034WM25 A0A0R1E6U2 B4PNE2 A0A1L8DSB6 A0A0Q9XEZ1 A0A0B4KI38 B4K8A4 Q9V9X4 A0A1L8DSH4 A0A0T6B099 A0A0R3NFW4 B4QSM8 B4I029 Q29BB3 B3P538 B4GYU1 A0A0Q9WFQ3 B4LWZ8 A0A023EPH1 Q16I17 A0A1W4UI24 A0A1B0AA10 B4JRX2 A0A2K7P6D8 J3JU25 A0A0M5J5J5 A0A2J7RE64 U4UMP6 A0A1B0BSJ1 A0A154PE20 A0A088A0V6 A0A067RM30 A0A182GDF3 A0A0L7QTG6 A0A1Q3F2S1 E0VD22 A0A2A3EMN7 A0A1B6E498 T1P8E0 B0X724 A0A0M8ZVG9 A0A2H8TPA2 A0A2S2N645 J9K017 D7EIW4 A0A2P8YD53 A0A2S2R147 A0A1Y1KS96 E2BR69 A0A1B6HUX9 A0A1B6FMN1 A0A182J706 A0A1B6M9X3 A0A084VI63 W5JII4 A0A2J7RE61 A0A182F7K2 A0A182QR25 A0A182NS67 A0A182YAM4 K7J7D2 F4WHD9 A0A182K5C6 A0A182VZ96 A0A232FET0 A0A158NBD7 A0A182R646 A0A195EWW1 A0A151X051 A0A182PGR2 A0A182L9N1 A0A182HLA4 Q7PKS9 A0A026X1N3 A0A151J4I9 A0A182WSF3 A0A195BRR2 A0A182VBY7 A0A1J1J4Z4
A0A0L0CRS8 A0A336M6Q7 D3TPX8 A0A1A9V7L8 A0A1A9YPY7 A0A1I8MNW6 A0A1I8P818 A0A0K8TZJ1 A0A0A1XPU2 W8ATZ5 B4NG41 A0A3B0JLB6 A0A3S2L2C5 A0A0P9C7L9 B3M098 A0A034WM25 A0A0R1E6U2 B4PNE2 A0A1L8DSB6 A0A0Q9XEZ1 A0A0B4KI38 B4K8A4 Q9V9X4 A0A1L8DSH4 A0A0T6B099 A0A0R3NFW4 B4QSM8 B4I029 Q29BB3 B3P538 B4GYU1 A0A0Q9WFQ3 B4LWZ8 A0A023EPH1 Q16I17 A0A1W4UI24 A0A1B0AA10 B4JRX2 A0A2K7P6D8 J3JU25 A0A0M5J5J5 A0A2J7RE64 U4UMP6 A0A1B0BSJ1 A0A154PE20 A0A088A0V6 A0A067RM30 A0A182GDF3 A0A0L7QTG6 A0A1Q3F2S1 E0VD22 A0A2A3EMN7 A0A1B6E498 T1P8E0 B0X724 A0A0M8ZVG9 A0A2H8TPA2 A0A2S2N645 J9K017 D7EIW4 A0A2P8YD53 A0A2S2R147 A0A1Y1KS96 E2BR69 A0A1B6HUX9 A0A1B6FMN1 A0A182J706 A0A1B6M9X3 A0A084VI63 W5JII4 A0A2J7RE61 A0A182F7K2 A0A182QR25 A0A182NS67 A0A182YAM4 K7J7D2 F4WHD9 A0A182K5C6 A0A182VZ96 A0A232FET0 A0A158NBD7 A0A182R646 A0A195EWW1 A0A151X051 A0A182PGR2 A0A182L9N1 A0A182HLA4 Q7PKS9 A0A026X1N3 A0A151J4I9 A0A182WSF3 A0A195BRR2 A0A182VBY7 A0A1J1J4Z4
EC Number
5.3.1.23
Pubmed
19121390
26354079
22118469
26108605
20353571
25315136
+ More
25830018 24495485 17994087 18057021 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15632085 24945155 17510324 22516182 23537049 24845553 26483478 20566863 18362917 19820115 29403074 28004739 20798317 24438588 20920257 23761445 25244985 20075255 21719571 28648823 21347285 20966253 12060762 12364791 24508170
25830018 24495485 17994087 18057021 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15632085 24945155 17510324 22516182 23537049 24845553 26483478 20566863 18362917 19820115 29403074 28004739 20798317 24438588 20920257 23761445 25244985 20075255 21719571 28648823 21347285 20966253 12060762 12364791 24508170
EMBL
BABH01010796
BABH01010797
DQ645455
KQ459984
KPJ18990.1
ODYU01000448
+ More
SOQ35247.1 AGBW02003014 OWR55593.1 KQ459220 KPJ02871.1 JRES01000007 KNC34917.1 UFQS01000635 UFQT01000635 SSX05615.1 SSX25974.1 EZ423480 ADD19756.1 GDHF01032618 GDHF01030048 GDHF01026946 GDHF01023845 GDHF01017108 GDHF01003068 JAI19696.1 JAI22266.1 JAI25368.1 JAI28469.1 JAI35206.1 JAI49246.1 GBXI01016862 GBXI01001320 JAC97429.1 JAD12972.1 GAMC01016873 JAB89682.1 CH964251 OUUW01000005 SPP81132.1 RSAL01000217 RVE44107.1 CH902617 KPU79541.1 GAKP01003273 GAKP01003272 JAC55679.1 CM000160 KRK04735.1 GFDF01004732 JAV09352.1 CH933806 KRG02163.1 AE014297 AGB96523.1 AY060664 GFDF01004733 JAV09351.1 LJIG01022454 KRT80510.1 CM000070 KRS99771.1 CM000364 CH480819 CH954182 CH479198 CH940650 KRF83436.1 GAPW01002211 JAC11387.1 CH478117 CH916373 APGK01039373 BT126737 KB740970 AEE61699.1 ENN76653.1 CP012526 ALC47844.1 NEVH01005277 PNF39120.1 KB632388 ERL94382.1 JXJN01019729 KQ434869 KZC09440.1 KK852427 KDR24083.1 JXUM01242290 KQ635445 KXJ62470.1 KQ414744 KOC61915.1 GFDL01013218 JAV21827.1 DS235070 EEB11278.1 KZ288208 PBC33025.1 GEDC01004532 JAS32766.1 KA644976 AFP59605.1 DS232433 EDS41718.1 KQ435859 KOX70443.1 GFXV01003667 MBW15472.1 GGMR01000025 MBY12644.1 ABLF02031390 KQ971380 EFA12349.2 PYGN01000690 PSN42191.1 GGMS01014548 MBY83751.1 GEZM01075242 GEZM01075241 JAV64184.1 GL449906 EFN81802.1 GECU01029230 JAS78476.1 GECZ01032443 GECZ01018312 JAS37326.1 JAS51457.1 GEBQ01007270 JAT32707.1 ATLV01013301 KE524850 KFB37657.1 ADMH02001150 ETN63911.1 PNF39121.1 AXCN02000267 GL888158 EGI66416.1 NNAY01000374 OXU28807.1 ADTU01010923 KQ981953 KYN32374.1 KQ982622 KYQ53715.1 APCN01000888 AJ439060 AAAB01008987 KK107039 EZA61921.1 KQ980151 KYN17519.1 KQ976417 KYM89705.1 CVRI01000067 CRL06534.1
SOQ35247.1 AGBW02003014 OWR55593.1 KQ459220 KPJ02871.1 JRES01000007 KNC34917.1 UFQS01000635 UFQT01000635 SSX05615.1 SSX25974.1 EZ423480 ADD19756.1 GDHF01032618 GDHF01030048 GDHF01026946 GDHF01023845 GDHF01017108 GDHF01003068 JAI19696.1 JAI22266.1 JAI25368.1 JAI28469.1 JAI35206.1 JAI49246.1 GBXI01016862 GBXI01001320 JAC97429.1 JAD12972.1 GAMC01016873 JAB89682.1 CH964251 OUUW01000005 SPP81132.1 RSAL01000217 RVE44107.1 CH902617 KPU79541.1 GAKP01003273 GAKP01003272 JAC55679.1 CM000160 KRK04735.1 GFDF01004732 JAV09352.1 CH933806 KRG02163.1 AE014297 AGB96523.1 AY060664 GFDF01004733 JAV09351.1 LJIG01022454 KRT80510.1 CM000070 KRS99771.1 CM000364 CH480819 CH954182 CH479198 CH940650 KRF83436.1 GAPW01002211 JAC11387.1 CH478117 CH916373 APGK01039373 BT126737 KB740970 AEE61699.1 ENN76653.1 CP012526 ALC47844.1 NEVH01005277 PNF39120.1 KB632388 ERL94382.1 JXJN01019729 KQ434869 KZC09440.1 KK852427 KDR24083.1 JXUM01242290 KQ635445 KXJ62470.1 KQ414744 KOC61915.1 GFDL01013218 JAV21827.1 DS235070 EEB11278.1 KZ288208 PBC33025.1 GEDC01004532 JAS32766.1 KA644976 AFP59605.1 DS232433 EDS41718.1 KQ435859 KOX70443.1 GFXV01003667 MBW15472.1 GGMR01000025 MBY12644.1 ABLF02031390 KQ971380 EFA12349.2 PYGN01000690 PSN42191.1 GGMS01014548 MBY83751.1 GEZM01075242 GEZM01075241 JAV64184.1 GL449906 EFN81802.1 GECU01029230 JAS78476.1 GECZ01032443 GECZ01018312 JAS37326.1 JAS51457.1 GEBQ01007270 JAT32707.1 ATLV01013301 KE524850 KFB37657.1 ADMH02001150 ETN63911.1 PNF39121.1 AXCN02000267 GL888158 EGI66416.1 NNAY01000374 OXU28807.1 ADTU01010923 KQ981953 KYN32374.1 KQ982622 KYQ53715.1 APCN01000888 AJ439060 AAAB01008987 KK107039 EZA61921.1 KQ980151 KYN17519.1 KQ976417 KYM89705.1 CVRI01000067 CRL06534.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000037069
UP000078200
+ More
UP000092443 UP000095301 UP000095300 UP000007798 UP000268350 UP000283053 UP000007801 UP000002282 UP000009192 UP000000803 UP000001819 UP000000304 UP000001292 UP000008711 UP000008744 UP000008792 UP000008820 UP000192221 UP000092445 UP000001070 UP000019118 UP000092553 UP000235965 UP000030742 UP000092460 UP000076502 UP000005203 UP000027135 UP000069940 UP000249989 UP000053825 UP000009046 UP000242457 UP000002320 UP000053105 UP000007819 UP000007266 UP000245037 UP000008237 UP000075880 UP000030765 UP000000673 UP000069272 UP000075886 UP000075884 UP000076408 UP000002358 UP000007755 UP000075881 UP000075920 UP000215335 UP000005205 UP000075900 UP000078541 UP000075809 UP000075885 UP000075882 UP000075840 UP000007062 UP000053097 UP000078492 UP000076407 UP000078540 UP000075903 UP000183832
UP000092443 UP000095301 UP000095300 UP000007798 UP000268350 UP000283053 UP000007801 UP000002282 UP000009192 UP000000803 UP000001819 UP000000304 UP000001292 UP000008711 UP000008744 UP000008792 UP000008820 UP000192221 UP000092445 UP000001070 UP000019118 UP000092553 UP000235965 UP000030742 UP000092460 UP000076502 UP000005203 UP000027135 UP000069940 UP000249989 UP000053825 UP000009046 UP000242457 UP000002320 UP000053105 UP000007819 UP000007266 UP000245037 UP000008237 UP000075880 UP000030765 UP000000673 UP000069272 UP000075886 UP000075884 UP000076408 UP000002358 UP000007755 UP000075881 UP000075920 UP000215335 UP000005205 UP000075900 UP000078541 UP000075809 UP000075885 UP000075882 UP000075840 UP000007062 UP000053097 UP000078492 UP000076407 UP000078540 UP000075903 UP000183832
Interpro
Gene 3D
ProteinModelPortal
H9IYZ3
Q0ZB81
A0A194RNT7
A0A2H1V342
A0A212FPF1
A0A194QHF3
+ More
A0A0L0CRS8 A0A336M6Q7 D3TPX8 A0A1A9V7L8 A0A1A9YPY7 A0A1I8MNW6 A0A1I8P818 A0A0K8TZJ1 A0A0A1XPU2 W8ATZ5 B4NG41 A0A3B0JLB6 A0A3S2L2C5 A0A0P9C7L9 B3M098 A0A034WM25 A0A0R1E6U2 B4PNE2 A0A1L8DSB6 A0A0Q9XEZ1 A0A0B4KI38 B4K8A4 Q9V9X4 A0A1L8DSH4 A0A0T6B099 A0A0R3NFW4 B4QSM8 B4I029 Q29BB3 B3P538 B4GYU1 A0A0Q9WFQ3 B4LWZ8 A0A023EPH1 Q16I17 A0A1W4UI24 A0A1B0AA10 B4JRX2 A0A2K7P6D8 J3JU25 A0A0M5J5J5 A0A2J7RE64 U4UMP6 A0A1B0BSJ1 A0A154PE20 A0A088A0V6 A0A067RM30 A0A182GDF3 A0A0L7QTG6 A0A1Q3F2S1 E0VD22 A0A2A3EMN7 A0A1B6E498 T1P8E0 B0X724 A0A0M8ZVG9 A0A2H8TPA2 A0A2S2N645 J9K017 D7EIW4 A0A2P8YD53 A0A2S2R147 A0A1Y1KS96 E2BR69 A0A1B6HUX9 A0A1B6FMN1 A0A182J706 A0A1B6M9X3 A0A084VI63 W5JII4 A0A2J7RE61 A0A182F7K2 A0A182QR25 A0A182NS67 A0A182YAM4 K7J7D2 F4WHD9 A0A182K5C6 A0A182VZ96 A0A232FET0 A0A158NBD7 A0A182R646 A0A195EWW1 A0A151X051 A0A182PGR2 A0A182L9N1 A0A182HLA4 Q7PKS9 A0A026X1N3 A0A151J4I9 A0A182WSF3 A0A195BRR2 A0A182VBY7 A0A1J1J4Z4
A0A0L0CRS8 A0A336M6Q7 D3TPX8 A0A1A9V7L8 A0A1A9YPY7 A0A1I8MNW6 A0A1I8P818 A0A0K8TZJ1 A0A0A1XPU2 W8ATZ5 B4NG41 A0A3B0JLB6 A0A3S2L2C5 A0A0P9C7L9 B3M098 A0A034WM25 A0A0R1E6U2 B4PNE2 A0A1L8DSB6 A0A0Q9XEZ1 A0A0B4KI38 B4K8A4 Q9V9X4 A0A1L8DSH4 A0A0T6B099 A0A0R3NFW4 B4QSM8 B4I029 Q29BB3 B3P538 B4GYU1 A0A0Q9WFQ3 B4LWZ8 A0A023EPH1 Q16I17 A0A1W4UI24 A0A1B0AA10 B4JRX2 A0A2K7P6D8 J3JU25 A0A0M5J5J5 A0A2J7RE64 U4UMP6 A0A1B0BSJ1 A0A154PE20 A0A088A0V6 A0A067RM30 A0A182GDF3 A0A0L7QTG6 A0A1Q3F2S1 E0VD22 A0A2A3EMN7 A0A1B6E498 T1P8E0 B0X724 A0A0M8ZVG9 A0A2H8TPA2 A0A2S2N645 J9K017 D7EIW4 A0A2P8YD53 A0A2S2R147 A0A1Y1KS96 E2BR69 A0A1B6HUX9 A0A1B6FMN1 A0A182J706 A0A1B6M9X3 A0A084VI63 W5JII4 A0A2J7RE61 A0A182F7K2 A0A182QR25 A0A182NS67 A0A182YAM4 K7J7D2 F4WHD9 A0A182K5C6 A0A182VZ96 A0A232FET0 A0A158NBD7 A0A182R646 A0A195EWW1 A0A151X051 A0A182PGR2 A0A182L9N1 A0A182HLA4 Q7PKS9 A0A026X1N3 A0A151J4I9 A0A182WSF3 A0A195BRR2 A0A182VBY7 A0A1J1J4Z4
PDB
4LDQ
E-value=7.48422e-107,
Score=989
Ontologies
PATHWAY
GO
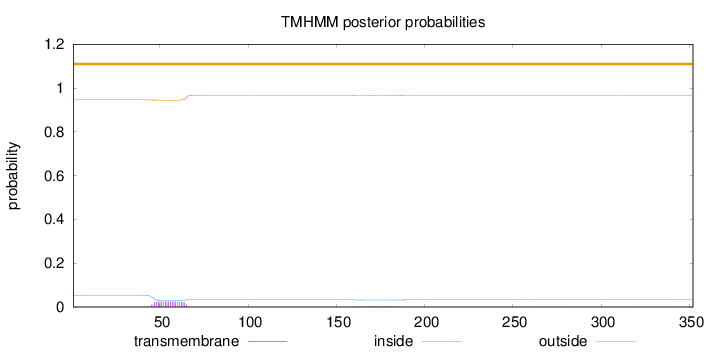
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
352
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.54409
Exp number, first 60 AAs:
0.3627
Total prob of N-in:
0.05360
outside
1 - 352
Population Genetic Test Statistics
Pi
221.550894
Theta
186.897087
Tajima's D
0.469108
CLR
0.070148
CSRT
0.501474926253687
Interpretation
Uncertain
