Gene
KWMTBOMO05228 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002429
Annotation
cyclophilin-like_protein_[Bombyx_mori]
Full name
Peptidyl-prolyl cis-trans isomerase
Alternative Name
Cyclophilin
Cyclosporin A-binding protein
Rotamase
Cyclosporin A-binding protein
Rotamase
Location in the cell
Cytoplasmic Reliability : 3.941
Sequence
CDS
ATGTCTCTACCACGAGTATTCTTCGACGTCACTGTTGACGATGCCCCACTGGGAAAAATTGTTATTGAGCTGAGAAGTGACGTCACTCCCAAGACGTGTGAAAACTTCCGTGCCCTGTGCACTGGCGAGAAAGGCTTCGGTTACAAGGGCTCCATTTTCCATCGTGTCATCCCCAATTTCATGCTGCAAGGAGGGGACTTCACCAACCATAACGGCACTGGGGGAAAGTCCATCTACGGCAATAAGTTTGAAGACGAGAATTTCACCCTTAAGCACACTGGACCTGGCGTCCTCTCCATGGCTAATGCCGGTGCTGATACTAATGGTTCCCAGTTCTTCATCACCACTGTCAAGACCTCCTGGCTGGATGGCAGACATGTTGTCTTTGGGAATGTTGTTGAAGGCATGGAAGTTGTCAAGCAGATTGAGACCTTTGGCAGCCAGTCTGGGAAGACCTCTAAGAGAATCGTTATCAAAGACTGTGGTCAGATTGCCTAA
Protein
MSLPRVFFDVTVDDAPLGKIVIELRSDVTPKTCENFRALCTGEKGFGYKGSIFHRVIPNFMLQGGDFTNHNGTGGKSIYGNKFEDENFTLKHTGPGVLSMANAGADTNGSQFFITTVKTSWLDGRHVVFGNVVEGMEVVKQIETFGSQSGKTSKRIVIKDCGQIA
Summary
Description
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Similarity
Belongs to the cyclophilin-type PPIase family.
Belongs to the cyclophilin-type PPIase family. PPIase A subfamily.
Belongs to the cyclophilin-type PPIase family. PPIase A subfamily.
Keywords
Complete proteome
Cytoplasm
Direct protein sequencing
Isomerase
Phosphoprotein
Reference proteome
Rotamase
Feature
chain Peptidyl-prolyl cis-trans isomerase
Uniprot
Q5CCL3
H9IYU4
Q56C53
I4DJA1
A0A3G1T1D0
I4DMT7
+ More
A0A2H1V328 R4FMV8 A0A194RNK0 A0A0L7KY60 S4PBG4 A0A0K8S607 A0A3S2NCR0 A0A212FPC4 U5EZE6 L7WSJ0 A0A182R0E6 D7RPB5 A0A2M4C0B2 A0A1Y1L1V2 A0A2M4BZN1 A0A1Y1L5L4 Q16P59 A0A2M3YY51 A0A2M4A133 A0A2M4A146 T1PHF9 T1DP12 A0A182IVW0 A0A023EJK1 A2I3U7 Q1HRR9 A0A0K8TQ86 A0A232FLD2 D6WB54 A0A182FMF3 A0A146LID2 A0A0T6AV63 A0A336LQB3 A0A182N180 A0A084VLS9 T1DGZ5 A0A1Z5KXX2 J9JT88 A0A182RHI0 A0A2R5LCA2 C4WRW7 A0A182IAP9 K7IM71 N6UJM4 A0A220XK87 A8C9W3 W5JUI0 B8RJ95 A0A182PFY1 B3GCD7 B3GCD6 A0A3S3SL31 A0A1D6T5K5 Q7PS16 A0A0P6IS29 A0A182LXJ5 A0A087T258 B7Q0J9 A0A2J7RN34 A1KXG2 J3JWH7 A0A1L8DU24 A0A0K8R8H4 A0A2H8TZ61 A0A1B6G2X9 A0A1L8E8G0 A0A1L8EF36 R4UW88 D3TMT7 A0A023FFI9 A0A1L8E917 B4L5M3 A0A291KZD7 K7J0P3 A0A1W7R9W4 A0A182XXS4 A0A1Y2HML4 A0A0M4ENB3 A0A023FXC1 A0A1I8PXH6 A0A1E1X7M1 A0A1L4AAU0 A0A023GF86 A0A139A349 J7HBB0 R4WCQ5 A0A182XDV5 B4IF57 A0A2A3EAB9 V9IIN8 A0A088A8D4 A8E774 P25007 A0A0A0VBV2
A0A2H1V328 R4FMV8 A0A194RNK0 A0A0L7KY60 S4PBG4 A0A0K8S607 A0A3S2NCR0 A0A212FPC4 U5EZE6 L7WSJ0 A0A182R0E6 D7RPB5 A0A2M4C0B2 A0A1Y1L1V2 A0A2M4BZN1 A0A1Y1L5L4 Q16P59 A0A2M3YY51 A0A2M4A133 A0A2M4A146 T1PHF9 T1DP12 A0A182IVW0 A0A023EJK1 A2I3U7 Q1HRR9 A0A0K8TQ86 A0A232FLD2 D6WB54 A0A182FMF3 A0A146LID2 A0A0T6AV63 A0A336LQB3 A0A182N180 A0A084VLS9 T1DGZ5 A0A1Z5KXX2 J9JT88 A0A182RHI0 A0A2R5LCA2 C4WRW7 A0A182IAP9 K7IM71 N6UJM4 A0A220XK87 A8C9W3 W5JUI0 B8RJ95 A0A182PFY1 B3GCD7 B3GCD6 A0A3S3SL31 A0A1D6T5K5 Q7PS16 A0A0P6IS29 A0A182LXJ5 A0A087T258 B7Q0J9 A0A2J7RN34 A1KXG2 J3JWH7 A0A1L8DU24 A0A0K8R8H4 A0A2H8TZ61 A0A1B6G2X9 A0A1L8E8G0 A0A1L8EF36 R4UW88 D3TMT7 A0A023FFI9 A0A1L8E917 B4L5M3 A0A291KZD7 K7J0P3 A0A1W7R9W4 A0A182XXS4 A0A1Y2HML4 A0A0M4ENB3 A0A023FXC1 A0A1I8PXH6 A0A1E1X7M1 A0A1L4AAU0 A0A023GF86 A0A139A349 J7HBB0 R4WCQ5 A0A182XDV5 B4IF57 A0A2A3EAB9 V9IIN8 A0A088A8D4 A8E774 P25007 A0A0A0VBV2
EC Number
5.2.1.8
Pubmed
19121390
22651552
26354079
26227816
23622113
22118469
+ More
23326229 28004739 17510324 25315136 24945155 26483478 17204158 26369729 28648823 18362917 19820115 26823975 24438588 24330624 28528879 20075255 23537049 17760985 20920257 23761445 18495616 12364791 14747013 17210077 22516182 20353571 17994087 28982170 25244985 28503490 27895657 25977457 23691247 10731132 12537572 1707759 8500545 18327897 24303891
23326229 28004739 17510324 25315136 24945155 26483478 17204158 26369729 28648823 18362917 19820115 26823975 24438588 24330624 28528879 20075255 23537049 17760985 20920257 23761445 18495616 12364791 14747013 17210077 22516182 20353571 17994087 28982170 25244985 28503490 27895657 25977457 23691247 10731132 12537572 1707759 8500545 18327897 24303891
EMBL
DQ201183
AB206402
ABA54173.1
BAD90848.1
BABH01010795
AY966877
+ More
AAX76988.1 AK401369 KQ459220 BAM17991.1 KPJ02870.1 MG846936 AXY94788.1 AK402605 BAM19227.1 ODYU01000448 SOQ35248.1 GAHY01000813 JAA76697.1 KQ459984 KPJ18989.1 JTDY01004473 KOB68105.1 GAIX01005922 JAA86638.1 GBRD01017243 JAG48584.1 RSAL01000217 RVE44108.1 AGBW02003014 OWR55594.1 GANO01000936 JAB58935.1 KC118982 AGC84404.1 AXCN02002156 HM113489 ADI58372.1 GGFJ01009287 MBW58428.1 GEZM01066883 JAV67669.1 GGFJ01009323 MBW58464.1 GEZM01066882 JAV67670.1 CH477793 EAT36139.1 GGFM01000451 MBW21202.1 GGFK01001141 MBW34462.1 GGFK01001144 MBW34465.1 KA647328 AFP61957.1 GAMD01002370 JAA99220.1 JXUM01086823 GAPW01004438 KQ563593 JAC09160.1 KXJ73620.1 EF070450 ABM55516.1 DQ440025 ABF18058.1 GDAI01001297 JAI16306.1 NNAY01000049 OXU31554.1 KQ971311 EEZ98965.2 GDHC01010788 JAQ07841.1 LJIG01022734 KRT78986.1 UFQT01000054 SSX19061.1 ATLV01014549 KE524974 KFB38923.1 GALA01001471 JAA93381.1 GFJQ02007329 JAV99640.1 ABLF02033823 GGLE01002982 MBY07108.1 AK339947 BAH70637.1 APCN01001299 APGK01021421 KB740322 KB631815 ENN80871.1 ERL86427.1 KY273928 ASL05640.1 EU032341 ABV44708.1 ADMH02000109 ETN67806.1 EZ000520 ACJ64394.1 EU669818 ACD69576.1 EU669817 ACD69575.1 NCKU01000192 RWS16668.1 AAAB01008846 EAA06299.6 GDIQ01002400 JAN92337.1 AXCM01003587 KK113043 KFM59197.1 ABJB010263896 DS832869 EEC12371.1 NEVH01002545 PNF42238.1 AY283280 KM010000 AAP35065.1 AIO08856.1 BT127595 AEE62557.1 GFDF01004141 JAV09943.1 GADI01006427 JAA67381.1 GFXV01007384 MBW19189.1 GECZ01012974 JAS56795.1 GFDG01003906 JAV14893.1 GFDG01001515 JAV17284.1 KC740692 AGM32516.1 EZ422739 ADD19015.1 GBBK01004617 JAC19865.1 GFDG01003700 JAV15099.1 CH933811 EDW06482.2 MF346049 ATI08942.1 GFAH01000450 JAV47939.1 MCFL01000020 ORZ35830.1 CP012526 ALC47077.1 GBBL01001965 JAC25355.1 GFAC01003935 JAT95253.1 KX268378 API65179.1 GBBM01002861 JAC32557.1 KQ965809 KXS11134.1 JX160103 AFP99100.1 AK416924 BAN20139.1 CH480832 EDW46111.1 KZ288325 PBC28011.1 JR049570 AEY61003.1 BT031016 ABV82398.1 AE014298 BT009946 M62398 KF860413 AIW62418.1
AAX76988.1 AK401369 KQ459220 BAM17991.1 KPJ02870.1 MG846936 AXY94788.1 AK402605 BAM19227.1 ODYU01000448 SOQ35248.1 GAHY01000813 JAA76697.1 KQ459984 KPJ18989.1 JTDY01004473 KOB68105.1 GAIX01005922 JAA86638.1 GBRD01017243 JAG48584.1 RSAL01000217 RVE44108.1 AGBW02003014 OWR55594.1 GANO01000936 JAB58935.1 KC118982 AGC84404.1 AXCN02002156 HM113489 ADI58372.1 GGFJ01009287 MBW58428.1 GEZM01066883 JAV67669.1 GGFJ01009323 MBW58464.1 GEZM01066882 JAV67670.1 CH477793 EAT36139.1 GGFM01000451 MBW21202.1 GGFK01001141 MBW34462.1 GGFK01001144 MBW34465.1 KA647328 AFP61957.1 GAMD01002370 JAA99220.1 JXUM01086823 GAPW01004438 KQ563593 JAC09160.1 KXJ73620.1 EF070450 ABM55516.1 DQ440025 ABF18058.1 GDAI01001297 JAI16306.1 NNAY01000049 OXU31554.1 KQ971311 EEZ98965.2 GDHC01010788 JAQ07841.1 LJIG01022734 KRT78986.1 UFQT01000054 SSX19061.1 ATLV01014549 KE524974 KFB38923.1 GALA01001471 JAA93381.1 GFJQ02007329 JAV99640.1 ABLF02033823 GGLE01002982 MBY07108.1 AK339947 BAH70637.1 APCN01001299 APGK01021421 KB740322 KB631815 ENN80871.1 ERL86427.1 KY273928 ASL05640.1 EU032341 ABV44708.1 ADMH02000109 ETN67806.1 EZ000520 ACJ64394.1 EU669818 ACD69576.1 EU669817 ACD69575.1 NCKU01000192 RWS16668.1 AAAB01008846 EAA06299.6 GDIQ01002400 JAN92337.1 AXCM01003587 KK113043 KFM59197.1 ABJB010263896 DS832869 EEC12371.1 NEVH01002545 PNF42238.1 AY283280 KM010000 AAP35065.1 AIO08856.1 BT127595 AEE62557.1 GFDF01004141 JAV09943.1 GADI01006427 JAA67381.1 GFXV01007384 MBW19189.1 GECZ01012974 JAS56795.1 GFDG01003906 JAV14893.1 GFDG01001515 JAV17284.1 KC740692 AGM32516.1 EZ422739 ADD19015.1 GBBK01004617 JAC19865.1 GFDG01003700 JAV15099.1 CH933811 EDW06482.2 MF346049 ATI08942.1 GFAH01000450 JAV47939.1 MCFL01000020 ORZ35830.1 CP012526 ALC47077.1 GBBL01001965 JAC25355.1 GFAC01003935 JAT95253.1 KX268378 API65179.1 GBBM01002861 JAC32557.1 KQ965809 KXS11134.1 JX160103 AFP99100.1 AK416924 BAN20139.1 CH480832 EDW46111.1 KZ288325 PBC28011.1 JR049570 AEY61003.1 BT031016 ABV82398.1 AE014298 BT009946 M62398 KF860413 AIW62418.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000283053
UP000007151
+ More
UP000075886 UP000008820 UP000095301 UP000075880 UP000069940 UP000249989 UP000215335 UP000007266 UP000069272 UP000075884 UP000030765 UP000007819 UP000075900 UP000075840 UP000002358 UP000019118 UP000030742 UP000000673 UP000075885 UP000285301 UP000007062 UP000075883 UP000054359 UP000001555 UP000235965 UP000009192 UP000076408 UP000193411 UP000092553 UP000095300 UP000070544 UP000076407 UP000001292 UP000242457 UP000005203 UP000000803
UP000075886 UP000008820 UP000095301 UP000075880 UP000069940 UP000249989 UP000215335 UP000007266 UP000069272 UP000075884 UP000030765 UP000007819 UP000075900 UP000075840 UP000002358 UP000019118 UP000030742 UP000000673 UP000075885 UP000285301 UP000007062 UP000075883 UP000054359 UP000001555 UP000235965 UP000009192 UP000076408 UP000193411 UP000092553 UP000095300 UP000070544 UP000076407 UP000001292 UP000242457 UP000005203 UP000000803
Pfam
PF00160 Pro_isomerase
Interpro
SUPFAM
SSF50891
SSF50891
Gene 3D
ProteinModelPortal
Q5CCL3
H9IYU4
Q56C53
I4DJA1
A0A3G1T1D0
I4DMT7
+ More
A0A2H1V328 R4FMV8 A0A194RNK0 A0A0L7KY60 S4PBG4 A0A0K8S607 A0A3S2NCR0 A0A212FPC4 U5EZE6 L7WSJ0 A0A182R0E6 D7RPB5 A0A2M4C0B2 A0A1Y1L1V2 A0A2M4BZN1 A0A1Y1L5L4 Q16P59 A0A2M3YY51 A0A2M4A133 A0A2M4A146 T1PHF9 T1DP12 A0A182IVW0 A0A023EJK1 A2I3U7 Q1HRR9 A0A0K8TQ86 A0A232FLD2 D6WB54 A0A182FMF3 A0A146LID2 A0A0T6AV63 A0A336LQB3 A0A182N180 A0A084VLS9 T1DGZ5 A0A1Z5KXX2 J9JT88 A0A182RHI0 A0A2R5LCA2 C4WRW7 A0A182IAP9 K7IM71 N6UJM4 A0A220XK87 A8C9W3 W5JUI0 B8RJ95 A0A182PFY1 B3GCD7 B3GCD6 A0A3S3SL31 A0A1D6T5K5 Q7PS16 A0A0P6IS29 A0A182LXJ5 A0A087T258 B7Q0J9 A0A2J7RN34 A1KXG2 J3JWH7 A0A1L8DU24 A0A0K8R8H4 A0A2H8TZ61 A0A1B6G2X9 A0A1L8E8G0 A0A1L8EF36 R4UW88 D3TMT7 A0A023FFI9 A0A1L8E917 B4L5M3 A0A291KZD7 K7J0P3 A0A1W7R9W4 A0A182XXS4 A0A1Y2HML4 A0A0M4ENB3 A0A023FXC1 A0A1I8PXH6 A0A1E1X7M1 A0A1L4AAU0 A0A023GF86 A0A139A349 J7HBB0 R4WCQ5 A0A182XDV5 B4IF57 A0A2A3EAB9 V9IIN8 A0A088A8D4 A8E774 P25007 A0A0A0VBV2
A0A2H1V328 R4FMV8 A0A194RNK0 A0A0L7KY60 S4PBG4 A0A0K8S607 A0A3S2NCR0 A0A212FPC4 U5EZE6 L7WSJ0 A0A182R0E6 D7RPB5 A0A2M4C0B2 A0A1Y1L1V2 A0A2M4BZN1 A0A1Y1L5L4 Q16P59 A0A2M3YY51 A0A2M4A133 A0A2M4A146 T1PHF9 T1DP12 A0A182IVW0 A0A023EJK1 A2I3U7 Q1HRR9 A0A0K8TQ86 A0A232FLD2 D6WB54 A0A182FMF3 A0A146LID2 A0A0T6AV63 A0A336LQB3 A0A182N180 A0A084VLS9 T1DGZ5 A0A1Z5KXX2 J9JT88 A0A182RHI0 A0A2R5LCA2 C4WRW7 A0A182IAP9 K7IM71 N6UJM4 A0A220XK87 A8C9W3 W5JUI0 B8RJ95 A0A182PFY1 B3GCD7 B3GCD6 A0A3S3SL31 A0A1D6T5K5 Q7PS16 A0A0P6IS29 A0A182LXJ5 A0A087T258 B7Q0J9 A0A2J7RN34 A1KXG2 J3JWH7 A0A1L8DU24 A0A0K8R8H4 A0A2H8TZ61 A0A1B6G2X9 A0A1L8E8G0 A0A1L8EF36 R4UW88 D3TMT7 A0A023FFI9 A0A1L8E917 B4L5M3 A0A291KZD7 K7J0P3 A0A1W7R9W4 A0A182XXS4 A0A1Y2HML4 A0A0M4ENB3 A0A023FXC1 A0A1I8PXH6 A0A1E1X7M1 A0A1L4AAU0 A0A023GF86 A0A139A349 J7HBB0 R4WCQ5 A0A182XDV5 B4IF57 A0A2A3EAB9 V9IIN8 A0A088A8D4 A8E774 P25007 A0A0A0VBV2
PDB
4TOT
E-value=2.04655e-70,
Score=670
Ontologies
GO
Topology
Subcellular location
Cytoplasm
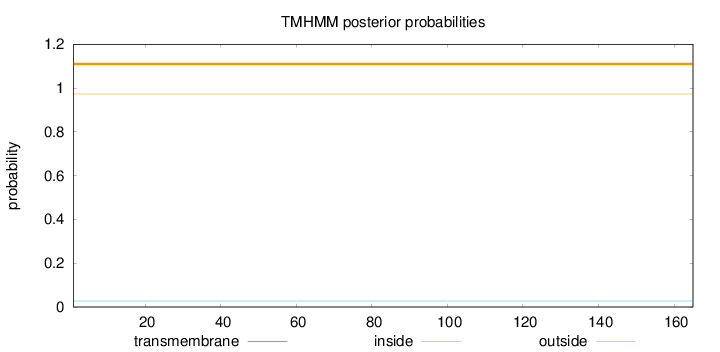
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00107
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.02697
outside
1 - 165
Population Genetic Test Statistics
Pi
157.401833
Theta
177.258642
Tajima's D
-0.542418
CLR
16.173452
CSRT
0.230688465576721
Interpretation
Uncertain
