Gene
KWMTBOMO05225
Pre Gene Modal
BGIBMGA002475
Annotation
A_disintegrin_and_metalloproteinase_with_thrombospondin_motifs_1_precursor_[Bombyx_mori]
Full name
A disintegrin and metalloproteinase with thrombospondin motifs 3
Alternative Name
Procollagen II N-proteinase
Procollagen II amino propeptide-processing enzyme
Procollagen II amino propeptide-processing enzyme
Location in the cell
Extracellular Reliability : 2.417 Nuclear Reliability : 2.394
Sequence
CDS
ATGCCTCACGTGTGCCGGTCAAAGCGAGCGCCGCCTCTCGAGGGAACACCCTGTGGACAAAACCAATGGTGCGTGGATCGTGTCTGCGAGCCGATGCCTGGCCACAGTAAAGAGACTAAGGTTGAAAACAAACATACTCCAGAATGGGGAGATTGGGAGGAGTGGAGCGCATGCAACGCAGATTGTGGTTACGGCCTCAGGACAAGAACACGAAAATGCAAATACAGAGGGTTTGTATCAGAGAGCGCATGTGAGGGTGCTGGATCTCAAGTAGCGACTTGCTGGGCGGGGTCTTCGTGCGCTGCAACCAGAGACATCCGCTCCGACCTCTGCCATCGCCAACAGAGCAGACTCATACCTTATTTACATGCCAACGAGTCCAACCATTGTGAAATTTCGTGTGTGGACTACGCTGGAGGGTCGCCCACCAATTTCGGAGCCCTACCCGACGGAACGCCCTGCAGCTACCTTCGACCCTTCGACGTTTGCTTCCAGGGTACGTGCGTGAAAGGTCAATGCAATTCATCAGATACAACCTGCAACTGGTGCCCAGACGGCTACTGCAACAACAACACTAACACATACACAAGATTACTCGGTAACGGCTGGACTCGCATGACGATGGTGCCGCACGAAGCACGTCAACTGTCCATACACATAGCCACACCGATTCCCCTCCACATTGCACTGCGCGAACGGAAACGGGACAAGCCGATTCTAGAACTTTCTAAGCACTCCAAAAAATTCGACATCAGCTCCCTTCAGGACAACTACTTGAAATACGATCCGAGCGTGCCTCAGAATCTTCAGATCGTGGAAATGGACAGCAACATTTTAGACTTGAAAGAGAGCTTTCGTTATGAAGGCGAGGCGATCACGGCTGGGACTCTCCTGCGCTGGAATCAAACCGACACGGACATTTATATAACGTCAGAATCTAGACTGCAAACTGATTTGATGATAATGGCAATACCAGTTAATCCTACATTAGAAGATGCGGTCAGTGTGGACGTGTCAGTAAACTACAGCACTCCAACTGGACGCACGAGACCATTGGAATACAGGTGGTCGATAGAGCGCGGTCCGTGTTCCGCGTCCTGCGGTGGCGGTGTGCGATTGATAACAGCCCAGTGTCATCGAGATCAGAAATGTCCACCGCCTAGATACGAATCATGCAATACACACAGCTGTGAATTTGTGTGGGCTTCTGACGATTGGGAAGAATGCAGTTCGACATGCGGTAGCAACGGTGTTCAAGAAAGACAATTGTTCTGTGTTCCATCAAACGCAAGCATGTTATCGAGGAGAGAGTTTATCAAACACAGCGTGTCACCAGTCATGTGCTCGTCGTCAAAGCCGCCACATCGCCAGCCCTGCAACCGTATCCCCTGCCCTGTTTACTGGAGGGAGCAACCCTGGACTCCTTGCTCGGCGTCGTGCGGACGCGGCGTGTCCCGGCGTCCGCTCACGTGTCCGGCGTCAGACGAGCTACTATGCGGACCGAAGCCGCGCGAGCGACGCCGGCGCTGCAGACTCCGGCGCTGCCCCTCAGCATTGAGAGTCGCAGTGCAGTGCCCCGAACGCGACCACGCGCACTACTGCGAGCTCTTCACACTGGAACAGCTGCACCGGAACTGCGAGGTGCCGCCGTTCAGGAAATATTGCTGTAACGCTTGCAGGGACGCAGACCGCCGTCAGCACAGACGGTACGGCTGA
Protein
MPHVCRSKRAPPLEGTPCGQNQWCVDRVCEPMPGHSKETKVENKHTPEWGDWEEWSACNADCGYGLRTRTRKCKYRGFVSESACEGAGSQVATCWAGSSCAATRDIRSDLCHRQQSRLIPYLHANESNHCEISCVDYAGGSPTNFGALPDGTPCSYLRPFDVCFQGTCVKGQCNSSDTTCNWCPDGYCNNNTNTYTRLLGNGWTRMTMVPHEARQLSIHIATPIPLHIALRERKRDKPILELSKHSKKFDISSLQDNYLKYDPSVPQNLQIVEMDSNILDLKESFRYEGEAITAGTLLRWNQTDTDIYITSESRLQTDLMIMAIPVNPTLEDAVSVDVSVNYSTPTGRTRPLEYRWSIERGPCSASCGGGVRLITAQCHRDQKCPPPRYESCNTHSCEFVWASDDWEECSSTCGSNGVQERQLFCVPSNASMLSRREFIKHSVSPVMCSSSKPPHRQPCNRIPCPVYWREQPWTPCSASCGRGVSRRPLTCPASDELLCGPKPRERRRRCRLRRCPSALRVAVQCPERDHAHYCELFTLEQLHRNCEVPPFRKYCCNACRDADRRQHRRYG
Summary
Cofactor
Zn(2+)
Keywords
Cleavage on pair of basic residues
Complete proteome
Disease mutation
Disulfide bond
Extracellular matrix
Glycoprotein
Heparin-binding
Hydrolase
Metal-binding
Metalloprotease
Polymorphism
Protease
Reference proteome
Repeat
Secreted
Signal
Zinc
Zymogen
Feature
chain A disintegrin and metalloproteinase with thrombospondin motifs 3
sequence variant In dbSNP:rs788908.
sequence variant In dbSNP:rs788908.
Uniprot
Q5W7F4
A0A2H1WAC8
A0A194RNT2
A0A194QHE7
S4NIS8
A0A2P8Z1U4
+ More
A0A2J7RN45 A0A067R067 A0A139WNG8 A0A091RHA2 A0A091KXR1 A0A091U841 A0A3B3WMY8 A0A3B3WMY7 A0A087XEN1 A0A2I0MH16 A0A093CHI1 A0A151N535 A0A091PZ28 A0A091P525 G1NCI6 A0A091H105 A0A091SXL7 A0A3Q0G275 A0A091K8M6 R0M107 U3IS63 A0A099ZNV3 A0A3P9PEY9 F1NVD1 A0A091VCW9 A0A091LC65 A0A3Q0EFX4 A0A087R272 D2I440 A0A091J468 G5ANH7 A0A2K5HK83 A0A093CCX3 A0A0N8ETJ2 A0A2U3YAU7 A0A0D9QXR4 G3WCC6 M3XK24 G3T767 A0A093FUL1 A0A2Y9I964 A0A3Q3WLY1 A0A2K6U7Q8 A0A3B3V2H4 A0A2K5YJI7 A0A2K6DXS9 A0A2K5VFV4 F7DSS6 F7A2S0 A0A2U3VE68 G7MT43 A0A3Q0T9Y4 A0A2K6KEF5 H2PDJ7 A0A096MW58 G1R882 A0A3B5QTX8 A0A3Q7Q9K0 A0A2K6PGE9 G3WCC5 M3ZJG0 A0A2J8UTV5 F6ZC90 O15072 M7BEC0 K7FWV3 A0A0A0APK9 A0A096N567 A0A2K5NRE9 A0A2K5NRF3 A0A2R9C2C1 A0A2I3TEV8 G7P5M2 A0A2R9CC15 A0A2K5DSM2 A0A2J8PFD7 A0A384C954 A0A3Q7VXP6 A0A287D421 A0A3B5LAW6 G3QLZ8 A0A2K5RWN8 F7EC85 H3ASK2 A0A3P9N0W4 A0A2Y9DZH5 A0A096MCV0 A0A3Q3ACX3 A0A3Q2PEV4 A0A0S7K3A9
A0A2J7RN45 A0A067R067 A0A139WNG8 A0A091RHA2 A0A091KXR1 A0A091U841 A0A3B3WMY8 A0A3B3WMY7 A0A087XEN1 A0A2I0MH16 A0A093CHI1 A0A151N535 A0A091PZ28 A0A091P525 G1NCI6 A0A091H105 A0A091SXL7 A0A3Q0G275 A0A091K8M6 R0M107 U3IS63 A0A099ZNV3 A0A3P9PEY9 F1NVD1 A0A091VCW9 A0A091LC65 A0A3Q0EFX4 A0A087R272 D2I440 A0A091J468 G5ANH7 A0A2K5HK83 A0A093CCX3 A0A0N8ETJ2 A0A2U3YAU7 A0A0D9QXR4 G3WCC6 M3XK24 G3T767 A0A093FUL1 A0A2Y9I964 A0A3Q3WLY1 A0A2K6U7Q8 A0A3B3V2H4 A0A2K5YJI7 A0A2K6DXS9 A0A2K5VFV4 F7DSS6 F7A2S0 A0A2U3VE68 G7MT43 A0A3Q0T9Y4 A0A2K6KEF5 H2PDJ7 A0A096MW58 G1R882 A0A3B5QTX8 A0A3Q7Q9K0 A0A2K6PGE9 G3WCC5 M3ZJG0 A0A2J8UTV5 F6ZC90 O15072 M7BEC0 K7FWV3 A0A0A0APK9 A0A096N567 A0A2K5NRE9 A0A2K5NRF3 A0A2R9C2C1 A0A2I3TEV8 G7P5M2 A0A2R9CC15 A0A2K5DSM2 A0A2J8PFD7 A0A384C954 A0A3Q7VXP6 A0A287D421 A0A3B5LAW6 G3QLZ8 A0A2K5RWN8 F7EC85 H3ASK2 A0A3P9N0W4 A0A2Y9DZH5 A0A096MCV0 A0A3Q3ACX3 A0A3Q2PEV4 A0A0S7K3A9
EC Number
3.4.24.-
Pubmed
15898116
26354079
23622113
29403074
24845553
18362917
+ More
19820115 23371554 22293439 20838655 23749191 15592404 20010809 21993625 21709235 9215903 17431167 19892987 22002653 23542700 25362486 15815621 15489334 11408482 9205841 28985353 23624526 17381049 22722832 16136131 24813606 22398555 17495919
19820115 23371554 22293439 20838655 23749191 15592404 20010809 21993625 21709235 9215903 17431167 19892987 22002653 23542700 25362486 15815621 15489334 11408482 9205841 28985353 23624526 17381049 22722832 16136131 24813606 22398555 17495919
EMBL
AB194270
BAD69559.2
ODYU01007276
SOQ49906.1
KQ459984
KPJ18985.1
+ More
KQ459220 KPJ02866.1 GAIX01013944 JAA78616.1 PYGN01000238 PSN50464.1 NEVH01002545 PNF42261.1 KK852834 KDR15277.1 KQ971311 KYB29417.1 KK799877 KFQ27987.1 KK758737 KFP44208.1 KK441088 KFQ70223.1 AYCK01002573 AYCK01002574 AYCK01002575 AYCK01002576 AYCK01002577 AKCR02000013 PKK28972.1 KL244624 KFV13923.1 AKHW03004011 KYO31943.1 KK652494 KFQ02619.1 KK668078 KFQ02695.1 KL448082 KFO80022.1 KK937324 KFQ47943.1 KK549618 KFP33380.1 KB742619 EOB06348.1 ADON01026404 ADON01026405 ADON01026406 KL896449 KGL83506.1 AADN05000017 KL410916 KFR00914.1 KL308792 KFP52695.1 KL226056 KFM07576.1 ACTA01106043 ACTA01114043 ACTA01122043 ACTA01130043 GL194421 EFB28993.1 KK501372 KFP14465.1 JH166213 EHA98587.1 KL458380 KFV10864.1 GEBF01004227 JAN99405.1 AQIB01016336 AQIB01016337 AQIB01016338 AQIB01016339 AQIB01016340 AQIB01016341 AEFK01211420 AEFK01211421 AEFK01211422 AEFK01211423 AEFK01211424 AEFK01211425 AEFK01211426 AEFK01211427 AEFK01211428 AEFK01211429 AFYH01085802 AFYH01085803 AFYH01085804 AFYH01085805 AFYH01085806 AFYH01085807 AFYH01085808 AFYH01085809 AFYH01085810 AFYH01085811 KK635260 KFV58056.1 AQIA01054839 AQIA01054840 AQIA01054841 AQIA01054842 AQIA01054843 AQIA01054844 AQIA01054845 AQIA01054846 AQIA01054847 AQIA01054848 JSUE03033558 JSUE03033559 JSUE03033560 CM001257 EHH25939.1 ABGA01228450 ABGA01228451 ABGA01228452 ABGA01228453 ABGA01368097 ABGA01368098 ABGA01368099 ABGA01368100 ABGA01368101 ABGA01368102 AHZZ02026886 AHZZ02026887 AHZZ02026888 AHZZ02026889 AHZZ02026890 AHZZ02026891 ADFV01029806 ADFV01029807 ADFV01029808 ADFV01029809 ADFV01029810 ADFV01029811 ADFV01029812 ADFV01029813 NDHI03003443 PNJ48693.1 PNJ48694.1 AC093790 AC095056 AC104814 BC130287 BC132735 AF247668 AB002364 KB549684 EMP30488.1 AGCU01102093 AGCU01102094 AGCU01102095 AGCU01102096 AGCU01102097 AGCU01102098 AGCU01102099 AGCU01102100 AGCU01102101 AGCU01102102 KL872293 KGL94995.1 AJFE02062842 AJFE02062843 AJFE02062844 AJFE02062845 AJFE02062846 AJFE02062847 AACZ04033876 AACZ04033877 AACZ04033878 AACZ04033879 AACZ04033880 CM001280 EHH53740.1 NBAG03000215 PNI82723.1 PNI82724.1 AGTP01072028 AGTP01072029 CABD030031336 CABD030031337 CABD030031338 CABD030031339 CABD030031340 CABD030031341 CABD030031342 CABD030031343 CABD030031344 CABD030031345 AYCK01007292 GBYX01223108 JAO71405.1
KQ459220 KPJ02866.1 GAIX01013944 JAA78616.1 PYGN01000238 PSN50464.1 NEVH01002545 PNF42261.1 KK852834 KDR15277.1 KQ971311 KYB29417.1 KK799877 KFQ27987.1 KK758737 KFP44208.1 KK441088 KFQ70223.1 AYCK01002573 AYCK01002574 AYCK01002575 AYCK01002576 AYCK01002577 AKCR02000013 PKK28972.1 KL244624 KFV13923.1 AKHW03004011 KYO31943.1 KK652494 KFQ02619.1 KK668078 KFQ02695.1 KL448082 KFO80022.1 KK937324 KFQ47943.1 KK549618 KFP33380.1 KB742619 EOB06348.1 ADON01026404 ADON01026405 ADON01026406 KL896449 KGL83506.1 AADN05000017 KL410916 KFR00914.1 KL308792 KFP52695.1 KL226056 KFM07576.1 ACTA01106043 ACTA01114043 ACTA01122043 ACTA01130043 GL194421 EFB28993.1 KK501372 KFP14465.1 JH166213 EHA98587.1 KL458380 KFV10864.1 GEBF01004227 JAN99405.1 AQIB01016336 AQIB01016337 AQIB01016338 AQIB01016339 AQIB01016340 AQIB01016341 AEFK01211420 AEFK01211421 AEFK01211422 AEFK01211423 AEFK01211424 AEFK01211425 AEFK01211426 AEFK01211427 AEFK01211428 AEFK01211429 AFYH01085802 AFYH01085803 AFYH01085804 AFYH01085805 AFYH01085806 AFYH01085807 AFYH01085808 AFYH01085809 AFYH01085810 AFYH01085811 KK635260 KFV58056.1 AQIA01054839 AQIA01054840 AQIA01054841 AQIA01054842 AQIA01054843 AQIA01054844 AQIA01054845 AQIA01054846 AQIA01054847 AQIA01054848 JSUE03033558 JSUE03033559 JSUE03033560 CM001257 EHH25939.1 ABGA01228450 ABGA01228451 ABGA01228452 ABGA01228453 ABGA01368097 ABGA01368098 ABGA01368099 ABGA01368100 ABGA01368101 ABGA01368102 AHZZ02026886 AHZZ02026887 AHZZ02026888 AHZZ02026889 AHZZ02026890 AHZZ02026891 ADFV01029806 ADFV01029807 ADFV01029808 ADFV01029809 ADFV01029810 ADFV01029811 ADFV01029812 ADFV01029813 NDHI03003443 PNJ48693.1 PNJ48694.1 AC093790 AC095056 AC104814 BC130287 BC132735 AF247668 AB002364 KB549684 EMP30488.1 AGCU01102093 AGCU01102094 AGCU01102095 AGCU01102096 AGCU01102097 AGCU01102098 AGCU01102099 AGCU01102100 AGCU01102101 AGCU01102102 KL872293 KGL94995.1 AJFE02062842 AJFE02062843 AJFE02062844 AJFE02062845 AJFE02062846 AJFE02062847 AACZ04033876 AACZ04033877 AACZ04033878 AACZ04033879 AACZ04033880 CM001280 EHH53740.1 NBAG03000215 PNI82723.1 PNI82724.1 AGTP01072028 AGTP01072029 CABD030031336 CABD030031337 CABD030031338 CABD030031339 CABD030031340 CABD030031341 CABD030031342 CABD030031343 CABD030031344 CABD030031345 AYCK01007292 GBYX01223108 JAO71405.1
Proteomes
UP000053240
UP000053268
UP000245037
UP000235965
UP000027135
UP000007266
+ More
UP000261480 UP000028760 UP000053872 UP000050525 UP000001645 UP000053760 UP000189705 UP000016666 UP000053641 UP000242638 UP000000539 UP000053283 UP000189704 UP000053286 UP000008912 UP000053119 UP000006813 UP000233080 UP000245341 UP000029965 UP000007648 UP000008672 UP000007646 UP000248481 UP000261620 UP000233220 UP000261500 UP000233140 UP000233120 UP000233100 UP000006718 UP000002281 UP000245340 UP000261340 UP000233180 UP000001595 UP000028761 UP000001073 UP000002852 UP000286641 UP000233200 UP000005640 UP000031443 UP000007267 UP000053858 UP000233060 UP000240080 UP000002277 UP000009130 UP000233020 UP000261680 UP000291021 UP000286642 UP000005215 UP000261380 UP000001519 UP000233040 UP000002280 UP000248480 UP000264800 UP000265000
UP000261480 UP000028760 UP000053872 UP000050525 UP000001645 UP000053760 UP000189705 UP000016666 UP000053641 UP000242638 UP000000539 UP000053283 UP000189704 UP000053286 UP000008912 UP000053119 UP000006813 UP000233080 UP000245341 UP000029965 UP000007648 UP000008672 UP000007646 UP000248481 UP000261620 UP000233220 UP000261500 UP000233140 UP000233120 UP000233100 UP000006718 UP000002281 UP000245340 UP000261340 UP000233180 UP000001595 UP000028761 UP000001073 UP000002852 UP000286641 UP000233200 UP000005640 UP000031443 UP000007267 UP000053858 UP000233060 UP000240080 UP000002277 UP000009130 UP000233020 UP000261680 UP000291021 UP000286642 UP000005215 UP000261380 UP000001519 UP000233040 UP000002280 UP000248480 UP000264800 UP000265000
Pfam
Interpro
SUPFAM
SSF82895
SSF82895
Gene 3D
ProteinModelPortal
Q5W7F4
A0A2H1WAC8
A0A194RNT2
A0A194QHE7
S4NIS8
A0A2P8Z1U4
+ More
A0A2J7RN45 A0A067R067 A0A139WNG8 A0A091RHA2 A0A091KXR1 A0A091U841 A0A3B3WMY8 A0A3B3WMY7 A0A087XEN1 A0A2I0MH16 A0A093CHI1 A0A151N535 A0A091PZ28 A0A091P525 G1NCI6 A0A091H105 A0A091SXL7 A0A3Q0G275 A0A091K8M6 R0M107 U3IS63 A0A099ZNV3 A0A3P9PEY9 F1NVD1 A0A091VCW9 A0A091LC65 A0A3Q0EFX4 A0A087R272 D2I440 A0A091J468 G5ANH7 A0A2K5HK83 A0A093CCX3 A0A0N8ETJ2 A0A2U3YAU7 A0A0D9QXR4 G3WCC6 M3XK24 G3T767 A0A093FUL1 A0A2Y9I964 A0A3Q3WLY1 A0A2K6U7Q8 A0A3B3V2H4 A0A2K5YJI7 A0A2K6DXS9 A0A2K5VFV4 F7DSS6 F7A2S0 A0A2U3VE68 G7MT43 A0A3Q0T9Y4 A0A2K6KEF5 H2PDJ7 A0A096MW58 G1R882 A0A3B5QTX8 A0A3Q7Q9K0 A0A2K6PGE9 G3WCC5 M3ZJG0 A0A2J8UTV5 F6ZC90 O15072 M7BEC0 K7FWV3 A0A0A0APK9 A0A096N567 A0A2K5NRE9 A0A2K5NRF3 A0A2R9C2C1 A0A2I3TEV8 G7P5M2 A0A2R9CC15 A0A2K5DSM2 A0A2J8PFD7 A0A384C954 A0A3Q7VXP6 A0A287D421 A0A3B5LAW6 G3QLZ8 A0A2K5RWN8 F7EC85 H3ASK2 A0A3P9N0W4 A0A2Y9DZH5 A0A096MCV0 A0A3Q3ACX3 A0A3Q2PEV4 A0A0S7K3A9
A0A2J7RN45 A0A067R067 A0A139WNG8 A0A091RHA2 A0A091KXR1 A0A091U841 A0A3B3WMY8 A0A3B3WMY7 A0A087XEN1 A0A2I0MH16 A0A093CHI1 A0A151N535 A0A091PZ28 A0A091P525 G1NCI6 A0A091H105 A0A091SXL7 A0A3Q0G275 A0A091K8M6 R0M107 U3IS63 A0A099ZNV3 A0A3P9PEY9 F1NVD1 A0A091VCW9 A0A091LC65 A0A3Q0EFX4 A0A087R272 D2I440 A0A091J468 G5ANH7 A0A2K5HK83 A0A093CCX3 A0A0N8ETJ2 A0A2U3YAU7 A0A0D9QXR4 G3WCC6 M3XK24 G3T767 A0A093FUL1 A0A2Y9I964 A0A3Q3WLY1 A0A2K6U7Q8 A0A3B3V2H4 A0A2K5YJI7 A0A2K6DXS9 A0A2K5VFV4 F7DSS6 F7A2S0 A0A2U3VE68 G7MT43 A0A3Q0T9Y4 A0A2K6KEF5 H2PDJ7 A0A096MW58 G1R882 A0A3B5QTX8 A0A3Q7Q9K0 A0A2K6PGE9 G3WCC5 M3ZJG0 A0A2J8UTV5 F6ZC90 O15072 M7BEC0 K7FWV3 A0A0A0APK9 A0A096N567 A0A2K5NRE9 A0A2K5NRF3 A0A2R9C2C1 A0A2I3TEV8 G7P5M2 A0A2R9CC15 A0A2K5DSM2 A0A2J8PFD7 A0A384C954 A0A3Q7VXP6 A0A287D421 A0A3B5LAW6 G3QLZ8 A0A2K5RWN8 F7EC85 H3ASK2 A0A3P9N0W4 A0A2Y9DZH5 A0A096MCV0 A0A3Q3ACX3 A0A3Q2PEV4 A0A0S7K3A9
PDB
2RJQ
E-value=1.10705e-05,
Score=119
Ontologies
GO
Topology
Subcellular location
Secreted
Extracellular space
Extracellular matrix
Extracellular space
Extracellular matrix
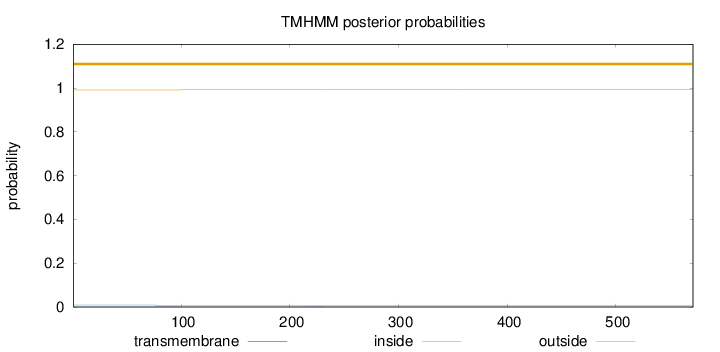
Length:
571
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01106
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00864
outside
1 - 571
Population Genetic Test Statistics
Pi
213.451511
Theta
166.538616
Tajima's D
0.781407
CLR
0.260426
CSRT
0.603869806509674
Interpretation
Uncertain
