Gene
KWMTBOMO05222
Pre Gene Modal
BGIBMGA002474
Annotation
A_disintegrin_and_metalloproteinase_with_thrombospondin_motifs_1_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.232 Mitochondrial Reliability : 1.398
Sequence
CDS
ATGCGTGAGCGCAGCGCGCCGCACCGCGACCCACAGAGAGGCGCAGCTCGCGGCGACGCACGGACCATGCCGTGCTGCCTCTGGGCCGCGCTGTCTCTGCTCCTCGCGGTCGTCGGCGCCGGCGCCTGGACACCGCCGAGACAGCCCTTGCTGCTCACTCCGAGACATCGCAGACACGCGCAAGAAACTCATCACTTGCGACTCCTCGGTTGGGATCTCAAGTTAAAAGAAAACAAAGCTATTCGTTCGCCTTATTATAAGGAATGTCAGTTTTTTACGGGACGCGTTCTTCATGAAGAGGGCTCCGCTGTGACGGTTACCGAATGTGACGGACAGCTGTACGGGCTGCTGCAGGTCGGCGGTGAAGAGTTCGTGCTGCAGCCGACCAGAACGCAGGAGAAACACGTGTTACGACGCCGAGACGTTACGCACAGCGAGCGTTCGGCCGAGTACAACCTCACCGGTGATACGGTCATCGACCTTGATCTTGACTTCGATGAAGATGATGACCTGTTACCGACCTCGCACGTACATCCGCGGCACAGCGATCATTCCGACAAAGAGTACTTCCATGACATGCATTTGTTCAGAAGACCTGCGTCAGGTGTAAAGGGTCTTTGGCTTGAACTGGCCATCGTTGCCGACAACACCATGTTGAAATTTCATGGCCGAGAACGTGTCAAACATTATATTTTAGCTTTAATGAATATTGTCAGTGCCATCTTCAATGACCCATCACTGGGCTCTAACATCACGTTAGTGATCAACAAATTGTTTCTCTACGAGGACAAGGACATAATTTTGAAGTACGGCAACATCAAAAAATCGTTAGAAGCTATTAACAAGTGGAACTATAGACATCTCATGAAGCTACCTGAAGGTAGCACAGGATGGGACGCCACAATATGGTTGACGAGATCCCAGCTTGGAGGGCCTTCCGGGTTCGCCCCCGTAGGTGGGGTCTGCACCAAAACGCGGTCAGCCGCCATCGACCGCGACGAAGGATTGACCAGTGCTTTTGTAATTGCACACGAACTCGCTCACTTATTAGGGTTGACCCACGATGGAGAAGGCAATTGTCAGTCTGAAGCGCTTCGAGGCTCTGTAATGGCACCGACAGTATTGGCTACCCTACACAACTTTGCCTGGTCAAGTTGTTCAAAGGAACAATTTCATGCGAAATCAAAGAAGTGGTGGTGCCTCCATGAGCGTAGCACTGACGAGGGTGTGGAACTAGGTGGTGCTAAGGAGCTCTCAAATTATGTCTTCACAATGGATGAACAGTGCAGGACTGAATTCGGAGAAGGGTAA
Protein
MRERSAPHRDPQRGAARGDARTMPCCLWAALSLLLAVVGAGAWTPPRQPLLLTPRHRRHAQETHHLRLLGWDLKLKENKAIRSPYYKECQFFTGRVLHEEGSAVTVTECDGQLYGLLQVGGEEFVLQPTRTQEKHVLRRRDVTHSERSAEYNLTGDTVIDLDLDFDEDDDLLPTSHVHPRHSDHSDKEYFHDMHLFRRPASGVKGLWLELAIVADNTMLKFHGRERVKHYILALMNIVSAIFNDPSLGSNITLVINKLFLYEDKDIILKYGNIKKSLEAINKWNYRHLMKLPEGSTGWDATIWLTRSQLGGPSGFAPVGGVCTKTRSAAIDRDEGLTSAFVIAHELAHLLGLTHDGEGNCQSEALRGSVMAPTVLATLHNFAWSSCSKEQFHAKSKKWWCLHERSTDEGVELGGAKELSNYVFTMDEQCRTEFGEG
Summary
Uniprot
Q5W7F4
H9IYY9
A0A3S2L1Y7
A0A194QHE7
A0A194RNT2
A0A2H1WA39
+ More
A0A232ESI1 A0A1W4WEI8 A0A1W4WEV2 A0A1W4WDK2 A0A088A8D9 A0A1Y1NAP5 A0A1Y1NDK9 A0A1Y1NAP7 A0A087ZT59 N6UKX4 A0A0M9A160 A0A139WN17 A0A139WNG8 A0A0C9RBA9 A0A2A3E8G8 A0A154PKF8 A0A0L7QZN0 T1IWV1 A0A0S7K1N5 A0A0Q3P5Y5 A0A1S3F9I3 A0A0S7K4W3 A0A151P062 A0A0S7K1K9 A0A3B4YNV9 A0A3P9N5L5 A0A3P9N0U9 A0A0S7K268 B2RXX5 A0A3B3C245 A0A3B5Q096 A0A3Q3IDJ4 A0A3Q0FV96 A0A1A7ZQ54 I3KGQ6 A0A3M0KF78 A0A3B4V8Y9 A0A3B4CC54 A0A3P8WFS5 M3ZWG6 A0A091P525 B2RXR8 A0A1A8IBL9 A0A3Q0T3H7 A0A3P8NSK8 A0A3B4YSZ2 A0A3Q0G275 A0A3P8TWY8 A0A091V4H7 A0A3Q1AYX9 A0A3B3V2H4 A0A096LXZ6 A0A091EU20 A0A094MUV0 A0A1W5AT79 A0A3S2P7Z4 A0A087XK85 W5UIH9 H2U8I4 A0A096MCV0 A0A3Q3MME4 A0A3B5LAW6 M4AL57 A0A0F8B1S6
A0A232ESI1 A0A1W4WEI8 A0A1W4WEV2 A0A1W4WDK2 A0A088A8D9 A0A1Y1NAP5 A0A1Y1NDK9 A0A1Y1NAP7 A0A087ZT59 N6UKX4 A0A0M9A160 A0A139WN17 A0A139WNG8 A0A0C9RBA9 A0A2A3E8G8 A0A154PKF8 A0A0L7QZN0 T1IWV1 A0A0S7K1N5 A0A0Q3P5Y5 A0A1S3F9I3 A0A0S7K4W3 A0A151P062 A0A0S7K1K9 A0A3B4YNV9 A0A3P9N5L5 A0A3P9N0U9 A0A0S7K268 B2RXX5 A0A3B3C245 A0A3B5Q096 A0A3Q3IDJ4 A0A3Q0FV96 A0A1A7ZQ54 I3KGQ6 A0A3M0KF78 A0A3B4V8Y9 A0A3B4CC54 A0A3P8WFS5 M3ZWG6 A0A091P525 B2RXR8 A0A1A8IBL9 A0A3Q0T3H7 A0A3P8NSK8 A0A3B4YSZ2 A0A3Q0G275 A0A3P8TWY8 A0A091V4H7 A0A3Q1AYX9 A0A3B3V2H4 A0A096LXZ6 A0A091EU20 A0A094MUV0 A0A1W5AT79 A0A3S2P7Z4 A0A087XK85 W5UIH9 H2U8I4 A0A096MCV0 A0A3Q3MME4 A0A3B5LAW6 M4AL57 A0A0F8B1S6
Pubmed
EMBL
AB194270
BAD69559.2
BABH01010769
RSAL01000231
RVE43838.1
KQ459220
+ More
KPJ02866.1 KQ459984 KPJ18985.1 ODYU01007276 SOQ49907.1 NNAY01002418 OXU21320.1 GEZM01007719 JAV94982.1 GEZM01007721 JAV94980.1 GEZM01007720 JAV94981.1 APGK01029202 KB740727 ENN79337.1 KQ435768 KOX75240.1 KQ971311 KYB29418.1 KYB29417.1 GBYB01005610 JAG75377.1 KZ288325 PBC28015.1 KQ434946 KZC12332.1 KQ414680 KOC64011.1 JH431630 GBYX01223105 JAO71408.1 LMAW01002878 KQK76163.1 GBYX01223110 JAO71403.1 AKHW03001467 KYO42424.1 GBYX01223117 GBYX01223109 JAO71404.1 GBYX01223116 JAO71397.1 AC123530 AC151895 BC158014 CH466553 AAI58015.1 EDL32150.1 HADY01005645 HAEJ01008758 SBP44130.1 AERX01007355 AERX01007356 QRBI01000108 RMC11848.1 KK668078 KFQ02695.1 BC157953 AAI57954.1 HAED01007483 SBQ93695.1 KL410604 KFQ97724.1 AYCK01006610 KK718970 KFO60491.1 KL350003 KFZ58286.1 CM012455 RVE59722.1 AYCK01007292 JT415790 AHH41821.1 KQ041942 KKF20668.1
KPJ02866.1 KQ459984 KPJ18985.1 ODYU01007276 SOQ49907.1 NNAY01002418 OXU21320.1 GEZM01007719 JAV94982.1 GEZM01007721 JAV94980.1 GEZM01007720 JAV94981.1 APGK01029202 KB740727 ENN79337.1 KQ435768 KOX75240.1 KQ971311 KYB29418.1 KYB29417.1 GBYB01005610 JAG75377.1 KZ288325 PBC28015.1 KQ434946 KZC12332.1 KQ414680 KOC64011.1 JH431630 GBYX01223105 JAO71408.1 LMAW01002878 KQK76163.1 GBYX01223110 JAO71403.1 AKHW03001467 KYO42424.1 GBYX01223117 GBYX01223109 JAO71404.1 GBYX01223116 JAO71397.1 AC123530 AC151895 BC158014 CH466553 AAI58015.1 EDL32150.1 HADY01005645 HAEJ01008758 SBP44130.1 AERX01007355 AERX01007356 QRBI01000108 RMC11848.1 KK668078 KFQ02695.1 BC157953 AAI57954.1 HAED01007483 SBQ93695.1 KL410604 KFQ97724.1 AYCK01006610 KK718970 KFO60491.1 KL350003 KFZ58286.1 CM012455 RVE59722.1 AYCK01007292 JT415790 AHH41821.1 KQ041942 KKF20668.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000215335
UP000192223
+ More
UP000005203 UP000019118 UP000053105 UP000007266 UP000242457 UP000076502 UP000053825 UP000051836 UP000081671 UP000050525 UP000261360 UP000242638 UP000000589 UP000261560 UP000002852 UP000261600 UP000189705 UP000005207 UP000269221 UP000261420 UP000261440 UP000265120 UP000261340 UP000265100 UP000265080 UP000053283 UP000257160 UP000261500 UP000028760 UP000052976 UP000192224 UP000005226 UP000261640 UP000261380
UP000005203 UP000019118 UP000053105 UP000007266 UP000242457 UP000076502 UP000053825 UP000051836 UP000081671 UP000050525 UP000261360 UP000242638 UP000000589 UP000261560 UP000002852 UP000261600 UP000189705 UP000005207 UP000269221 UP000261420 UP000261440 UP000265120 UP000261340 UP000265100 UP000265080 UP000053283 UP000257160 UP000261500 UP000028760 UP000052976 UP000192224 UP000005226 UP000261640 UP000261380
Pfam
Interpro
SUPFAM
SSF82895
SSF82895
Gene 3D
ProteinModelPortal
Q5W7F4
H9IYY9
A0A3S2L1Y7
A0A194QHE7
A0A194RNT2
A0A2H1WA39
+ More
A0A232ESI1 A0A1W4WEI8 A0A1W4WEV2 A0A1W4WDK2 A0A088A8D9 A0A1Y1NAP5 A0A1Y1NDK9 A0A1Y1NAP7 A0A087ZT59 N6UKX4 A0A0M9A160 A0A139WN17 A0A139WNG8 A0A0C9RBA9 A0A2A3E8G8 A0A154PKF8 A0A0L7QZN0 T1IWV1 A0A0S7K1N5 A0A0Q3P5Y5 A0A1S3F9I3 A0A0S7K4W3 A0A151P062 A0A0S7K1K9 A0A3B4YNV9 A0A3P9N5L5 A0A3P9N0U9 A0A0S7K268 B2RXX5 A0A3B3C245 A0A3B5Q096 A0A3Q3IDJ4 A0A3Q0FV96 A0A1A7ZQ54 I3KGQ6 A0A3M0KF78 A0A3B4V8Y9 A0A3B4CC54 A0A3P8WFS5 M3ZWG6 A0A091P525 B2RXR8 A0A1A8IBL9 A0A3Q0T3H7 A0A3P8NSK8 A0A3B4YSZ2 A0A3Q0G275 A0A3P8TWY8 A0A091V4H7 A0A3Q1AYX9 A0A3B3V2H4 A0A096LXZ6 A0A091EU20 A0A094MUV0 A0A1W5AT79 A0A3S2P7Z4 A0A087XK85 W5UIH9 H2U8I4 A0A096MCV0 A0A3Q3MME4 A0A3B5LAW6 M4AL57 A0A0F8B1S6
A0A232ESI1 A0A1W4WEI8 A0A1W4WEV2 A0A1W4WDK2 A0A088A8D9 A0A1Y1NAP5 A0A1Y1NDK9 A0A1Y1NAP7 A0A087ZT59 N6UKX4 A0A0M9A160 A0A139WN17 A0A139WNG8 A0A0C9RBA9 A0A2A3E8G8 A0A154PKF8 A0A0L7QZN0 T1IWV1 A0A0S7K1N5 A0A0Q3P5Y5 A0A1S3F9I3 A0A0S7K4W3 A0A151P062 A0A0S7K1K9 A0A3B4YNV9 A0A3P9N5L5 A0A3P9N0U9 A0A0S7K268 B2RXX5 A0A3B3C245 A0A3B5Q096 A0A3Q3IDJ4 A0A3Q0FV96 A0A1A7ZQ54 I3KGQ6 A0A3M0KF78 A0A3B4V8Y9 A0A3B4CC54 A0A3P8WFS5 M3ZWG6 A0A091P525 B2RXR8 A0A1A8IBL9 A0A3Q0T3H7 A0A3P8NSK8 A0A3B4YSZ2 A0A3Q0G275 A0A3P8TWY8 A0A091V4H7 A0A3Q1AYX9 A0A3B3V2H4 A0A096LXZ6 A0A091EU20 A0A094MUV0 A0A1W5AT79 A0A3S2P7Z4 A0A087XK85 W5UIH9 H2U8I4 A0A096MCV0 A0A3Q3MME4 A0A3B5LAW6 M4AL57 A0A0F8B1S6
PDB
2V4B
E-value=4.35624e-21,
Score=250
Ontologies
GO
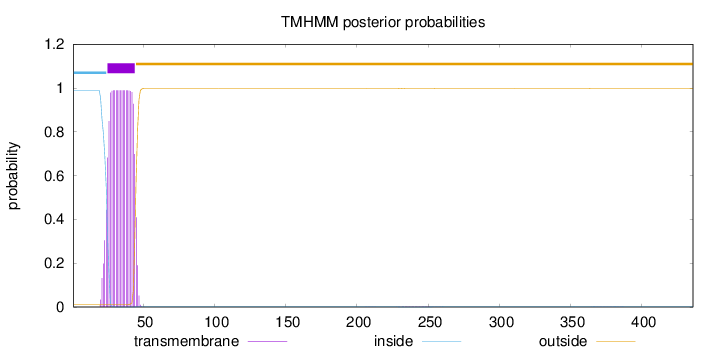
Topology
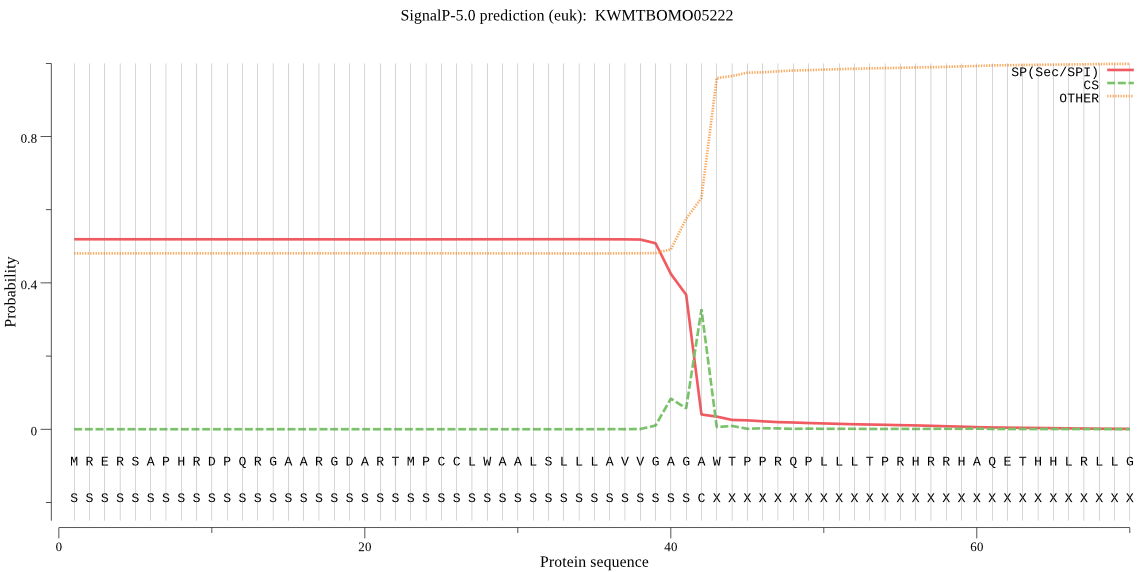
SignalP
Position: 1 - 42,
Likelihood: 0.519495
Length:
436
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.79601
Exp number, first 60 AAs:
20.7429
Total prob of N-in:
0.98991
POSSIBLE N-term signal
sequence
inside
1 - 24
TMhelix
25 - 44
outside
45 - 436
Population Genetic Test Statistics
Pi
187.527411
Theta
170.213631
Tajima's D
0.209719
CLR
0.106331
CSRT
0.432328383580821
Interpretation
Uncertain
