Gene
KWMTBOMO05217
Pre Gene Modal
BGIBMGA002471
Annotation
PREDICTED:_proton-associated_sugar_transporter_A-like_isoform_X2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.054
Sequence
CDS
ATGGCTGATAAACTTCATGAGTACCAGGGGTTAACAGGAAGGTTTCATAGTGCACGGGACCGATACAAAGACAGATGGTCAACATGGAAGGAGCAACATCCTAGGGGAGTCAAGGGCATCTTAGCGGAGACCTTGTTTGGTATCCCTAATTCAGAAGAGCCTTCTGGACGAGTATGGCAGGATAGCGAATATAGTGATATATTTAGGAGGAAAAGCCGCGTCGAGTTAATGCGTATTTCTGCCGCCGTAATGGGCATAGAATTCTCATACGCCGGCGAAACCGCTTTCGTGTCTCCGACGTTACTACAGATTGGGGTACCGCATGAAGAAATGACTTTAGTATGGGCTCTGTCACCGTTGATAGGGTTCTTCATGACTCCACTACTGGGTTCATTGAGTGACAGATGTCGTTCGAAGTTTGGGAGACGCAGGCCGTTTATAGTTTTACTATCGTTGGGCGTTTTTCTAGGTTTAATTCTAGTGCCTAACGGAGAGTCAATAGGCTACCTGTTAGGCGACGAGTATTCATCGAATTCGACTTCAACGCCCGCCGTGCTAGGACCCCGCAGTTCACTGGAAACTCCAGAAAAGAACTATCATCCCTGGGGCGTAGTGTTCACAGTTTTAGGAACTGTGTTCTTGGATTTCGACGCAGACGCCTGTCAGAGTCCAGCCCGGGCTTATCTCCTCGACGTGACAGTACCAGAGGATCATGCAAAAGGACTAAGTACTTTTACGGTGATGGCCGGCCTGGGAGGTTTTATGGGATACGCACTCGGAGGCATCAACTGGGACGAAACTAAACTAGGTATTTAA
Protein
MADKLHEYQGLTGRFHSARDRYKDRWSTWKEQHPRGVKGILAETLFGIPNSEEPSGRVWQDSEYSDIFRRKSRVELMRISAAVMGIEFSYAGETAFVSPTLLQIGVPHEEMTLVWALSPLIGFFMTPLLGSLSDRCRSKFGRRRPFIVLLSLGVFLGLILVPNGESIGYLLGDEYSSNSTSTPAVLGPRSSLETPEKNYHPWGVVFTVLGTVFLDFDADACQSPARAYLLDVTVPEDHAKGLSTFTVMAGLGGFMGYALGGINWDETKLGI
Summary
Uniprot
H9IYY6
A0A2H1VZC1
A0A3S2TEK5
A0A2A4IU42
A0A212ESH6
A0A194RN92
+ More
A0A194QBR9 A0A1W4WRJ1 A0A158NM21 A0A195CX99 A0A087ZQY3 F4WHS9 A0A0R3NMZ0 A0A067RHI5 A0A195BQV5 D6WAI6 A0A0M8ZQL2 B4N968 A0A026WEV7 A0A310SHN9 A0A151X0P3 A0A3B0KAZ0 A0A151JPZ4 E2B478 A0A0L0CME5 B5DWP1 A0A182U1N1 A0A182SQ16 A0A182WY99 A0A182VF87 A0A182HIH2 A0A1I8NMM5 A0A151JVU1 Q7QET1 U4U3Y9 N6UAV2 A0A182Q0W9 A0A0R1EE59 A0A336M410 A0A232F927 A0A0P8XWI3 A0A182YR91 A0A1B0B2J7 A0A1A9VFU1 A0A1A9YT10 B4K4F8 A0A1B0FI81 A0A224XLW9 T1HA98 A0A182MDE8 A0A182RL51 A0A182PV64 A0A0Q5WCU9 A0A0R1EBS8 A0A182W439 A0A182KAD8 A0A0R1E825 A0A0S0WGX2 A0A1B6JA19 A0A1B6HCC0 W8C3Y7 A0A023F2V7 A0A1B6D576 A0A1B6EXA3 Q6NL41 A0A154PMZ9 A0A0L7RCF6 A0A2R7W199 A0A182IM49 A0A1S4JX30 E1ZV04 B4MC78 A0A0J7KH91 A0A182FMD6 A0A084VV02 E0W3L9 A0A1Q3FRP6 A0A2H8TJK6 A0A1J1HRG4 A0A2S2R3J0 A0A0K8ULM7 A0A0A1WQY2 A0A2S2NKQ7 A0A1A9WYM2 A0A1I8NID2 A0A0P5NBE9 A0A0P5NUT5 A0A0P4YKL2 A0A162T655 A0A182NAG7 A0A0P4ZAN5 A0A0P4XXB4 A0A0N8DEH6 A0A0P5YEQ8 A0A0P5LRD3 A0A0P5VS04 A0A0K8T4S3 A0A146LVX5 A0A0A9YIN9 A0A0P5ZI78
A0A194QBR9 A0A1W4WRJ1 A0A158NM21 A0A195CX99 A0A087ZQY3 F4WHS9 A0A0R3NMZ0 A0A067RHI5 A0A195BQV5 D6WAI6 A0A0M8ZQL2 B4N968 A0A026WEV7 A0A310SHN9 A0A151X0P3 A0A3B0KAZ0 A0A151JPZ4 E2B478 A0A0L0CME5 B5DWP1 A0A182U1N1 A0A182SQ16 A0A182WY99 A0A182VF87 A0A182HIH2 A0A1I8NMM5 A0A151JVU1 Q7QET1 U4U3Y9 N6UAV2 A0A182Q0W9 A0A0R1EE59 A0A336M410 A0A232F927 A0A0P8XWI3 A0A182YR91 A0A1B0B2J7 A0A1A9VFU1 A0A1A9YT10 B4K4F8 A0A1B0FI81 A0A224XLW9 T1HA98 A0A182MDE8 A0A182RL51 A0A182PV64 A0A0Q5WCU9 A0A0R1EBS8 A0A182W439 A0A182KAD8 A0A0R1E825 A0A0S0WGX2 A0A1B6JA19 A0A1B6HCC0 W8C3Y7 A0A023F2V7 A0A1B6D576 A0A1B6EXA3 Q6NL41 A0A154PMZ9 A0A0L7RCF6 A0A2R7W199 A0A182IM49 A0A1S4JX30 E1ZV04 B4MC78 A0A0J7KH91 A0A182FMD6 A0A084VV02 E0W3L9 A0A1Q3FRP6 A0A2H8TJK6 A0A1J1HRG4 A0A2S2R3J0 A0A0K8ULM7 A0A0A1WQY2 A0A2S2NKQ7 A0A1A9WYM2 A0A1I8NID2 A0A0P5NBE9 A0A0P5NUT5 A0A0P4YKL2 A0A162T655 A0A182NAG7 A0A0P4ZAN5 A0A0P4XXB4 A0A0N8DEH6 A0A0P5YEQ8 A0A0P5LRD3 A0A0P5VS04 A0A0K8T4S3 A0A146LVX5 A0A0A9YIN9 A0A0P5ZI78
Pubmed
19121390
22118469
26354079
21347285
21719571
15632085
+ More
17994087 24845553 18362917 19820115 24508170 20798317 26108605 12364791 14747013 17210077 23537049 17550304 28648823 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 25474469 24438588 20566863 25830018 25315136 26823975 25401762
17994087 24845553 18362917 19820115 24508170 20798317 26108605 12364791 14747013 17210077 23537049 17550304 28648823 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 25474469 24438588 20566863 25830018 25315136 26823975 25401762
EMBL
BABH01010764
ODYU01005391
SOQ46181.1
RSAL01000231
RVE43841.1
NWSH01008211
+ More
PCG62650.1 AGBW02012778 OWR44448.1 KQ459984 KPJ18982.1 KQ459220 KPJ02864.1 ADTU01020148 ADTU01020149 KQ977171 KYN05177.1 GL888167 EGI66286.1 CM000070 KRT00605.1 KK852505 KDR22503.1 KQ976421 KYM89116.1 KQ971312 EEZ98637.2 KQ435911 KOX68927.1 CH964232 EDW81615.2 KK107238 EZA54615.1 KQ769179 OAD52952.1 KQ982610 KYQ53910.1 OUUW01000007 SPP83269.1 KQ978684 KYN29303.1 GL445515 EFN89495.1 JRES01000189 KNC33470.1 EDY68183.3 APCN01003970 KQ981688 KYN37930.1 AAAB01008846 EAA06394.5 KB631903 ERL87048.1 APGK01042213 APGK01042214 KB741002 ENN75762.1 AXCN02002022 CH892194 KRK05476.1 UFQT01000488 SSX24710.1 NNAY01000620 OXU27354.1 CH902618 KPU79127.1 JXJN01007683 CH933806 EDW13910.2 CCAG010001331 GFTR01007417 JAW09009.1 ACPB03019799 AXCM01000646 CH954177 KQS71085.1 KRK05475.1 KRK05474.1 KRK05478.1 AE014296 ALI30498.1 GECU01011635 JAS96071.1 GECU01035454 JAS72252.1 GAMC01002577 JAC03979.1 GBBI01003037 JAC15675.1 GEDC01016454 JAS20844.1 GECZ01027160 JAS42609.1 BT012500 AAS93771.1 KQ434960 KZC12678.1 KQ414616 KOC68421.1 KK854234 PTY13513.1 GL434335 EFN75002.1 CH940657 EDW63160.2 LBMM01007623 KMQ89571.1 ATLV01017139 KE525157 KFB41796.1 DS235882 EEB20225.1 GFDL01004775 JAV30270.1 GFXV01002528 MBW14333.1 CVRI01000020 CRK90613.1 GGMS01015398 MBY84601.1 GDHF01024712 JAI27602.1 GBXI01013372 JAD00920.1 GGMR01005161 MBY17780.1 GDIQ01147508 JAL04218.1 GDIQ01137421 JAL14305.1 GDIP01226075 JAI97326.1 LRGB01000007 KZS21943.1 GDIP01219570 JAJ03832.1 GDIP01235428 JAI87973.1 GDIP01041639 JAM62076.1 GDIP01058901 JAM44814.1 GDIQ01168361 JAK83364.1 GDIP01096126 JAM07589.1 GBRD01005304 JAG60517.1 GDHC01006805 JAQ11824.1 GBHO01011575 JAG32029.1 GDIP01044837 JAM58878.1
PCG62650.1 AGBW02012778 OWR44448.1 KQ459984 KPJ18982.1 KQ459220 KPJ02864.1 ADTU01020148 ADTU01020149 KQ977171 KYN05177.1 GL888167 EGI66286.1 CM000070 KRT00605.1 KK852505 KDR22503.1 KQ976421 KYM89116.1 KQ971312 EEZ98637.2 KQ435911 KOX68927.1 CH964232 EDW81615.2 KK107238 EZA54615.1 KQ769179 OAD52952.1 KQ982610 KYQ53910.1 OUUW01000007 SPP83269.1 KQ978684 KYN29303.1 GL445515 EFN89495.1 JRES01000189 KNC33470.1 EDY68183.3 APCN01003970 KQ981688 KYN37930.1 AAAB01008846 EAA06394.5 KB631903 ERL87048.1 APGK01042213 APGK01042214 KB741002 ENN75762.1 AXCN02002022 CH892194 KRK05476.1 UFQT01000488 SSX24710.1 NNAY01000620 OXU27354.1 CH902618 KPU79127.1 JXJN01007683 CH933806 EDW13910.2 CCAG010001331 GFTR01007417 JAW09009.1 ACPB03019799 AXCM01000646 CH954177 KQS71085.1 KRK05475.1 KRK05474.1 KRK05478.1 AE014296 ALI30498.1 GECU01011635 JAS96071.1 GECU01035454 JAS72252.1 GAMC01002577 JAC03979.1 GBBI01003037 JAC15675.1 GEDC01016454 JAS20844.1 GECZ01027160 JAS42609.1 BT012500 AAS93771.1 KQ434960 KZC12678.1 KQ414616 KOC68421.1 KK854234 PTY13513.1 GL434335 EFN75002.1 CH940657 EDW63160.2 LBMM01007623 KMQ89571.1 ATLV01017139 KE525157 KFB41796.1 DS235882 EEB20225.1 GFDL01004775 JAV30270.1 GFXV01002528 MBW14333.1 CVRI01000020 CRK90613.1 GGMS01015398 MBY84601.1 GDHF01024712 JAI27602.1 GBXI01013372 JAD00920.1 GGMR01005161 MBY17780.1 GDIQ01147508 JAL04218.1 GDIQ01137421 JAL14305.1 GDIP01226075 JAI97326.1 LRGB01000007 KZS21943.1 GDIP01219570 JAJ03832.1 GDIP01235428 JAI87973.1 GDIP01041639 JAM62076.1 GDIP01058901 JAM44814.1 GDIQ01168361 JAK83364.1 GDIP01096126 JAM07589.1 GBRD01005304 JAG60517.1 GDHC01006805 JAQ11824.1 GBHO01011575 JAG32029.1 GDIP01044837 JAM58878.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000192223 UP000005205 UP000078542 UP000005203 UP000007755 UP000001819 UP000027135 UP000078540 UP000007266 UP000053105 UP000007798 UP000053097 UP000075809 UP000268350 UP000078492 UP000008237 UP000037069 UP000075902 UP000075901 UP000076407 UP000075903 UP000075840 UP000095300 UP000078541 UP000007062 UP000030742 UP000019118 UP000075886 UP000002282 UP000215335 UP000007801 UP000076408 UP000092460 UP000078200 UP000092443 UP000009192 UP000092444 UP000015103 UP000075883 UP000075900 UP000075885 UP000008711 UP000075920 UP000075881 UP000000803 UP000076502 UP000053825 UP000075880 UP000000311 UP000008792 UP000036403 UP000069272 UP000030765 UP000009046 UP000183832 UP000091820 UP000095301 UP000076858 UP000075884
UP000192223 UP000005205 UP000078542 UP000005203 UP000007755 UP000001819 UP000027135 UP000078540 UP000007266 UP000053105 UP000007798 UP000053097 UP000075809 UP000268350 UP000078492 UP000008237 UP000037069 UP000075902 UP000075901 UP000076407 UP000075903 UP000075840 UP000095300 UP000078541 UP000007062 UP000030742 UP000019118 UP000075886 UP000002282 UP000215335 UP000007801 UP000076408 UP000092460 UP000078200 UP000092443 UP000009192 UP000092444 UP000015103 UP000075883 UP000075900 UP000075885 UP000008711 UP000075920 UP000075881 UP000000803 UP000076502 UP000053825 UP000075880 UP000000311 UP000008792 UP000036403 UP000069272 UP000030765 UP000009046 UP000183832 UP000091820 UP000095301 UP000076858 UP000075884
Interpro
CDD
ProteinModelPortal
H9IYY6
A0A2H1VZC1
A0A3S2TEK5
A0A2A4IU42
A0A212ESH6
A0A194RN92
+ More
A0A194QBR9 A0A1W4WRJ1 A0A158NM21 A0A195CX99 A0A087ZQY3 F4WHS9 A0A0R3NMZ0 A0A067RHI5 A0A195BQV5 D6WAI6 A0A0M8ZQL2 B4N968 A0A026WEV7 A0A310SHN9 A0A151X0P3 A0A3B0KAZ0 A0A151JPZ4 E2B478 A0A0L0CME5 B5DWP1 A0A182U1N1 A0A182SQ16 A0A182WY99 A0A182VF87 A0A182HIH2 A0A1I8NMM5 A0A151JVU1 Q7QET1 U4U3Y9 N6UAV2 A0A182Q0W9 A0A0R1EE59 A0A336M410 A0A232F927 A0A0P8XWI3 A0A182YR91 A0A1B0B2J7 A0A1A9VFU1 A0A1A9YT10 B4K4F8 A0A1B0FI81 A0A224XLW9 T1HA98 A0A182MDE8 A0A182RL51 A0A182PV64 A0A0Q5WCU9 A0A0R1EBS8 A0A182W439 A0A182KAD8 A0A0R1E825 A0A0S0WGX2 A0A1B6JA19 A0A1B6HCC0 W8C3Y7 A0A023F2V7 A0A1B6D576 A0A1B6EXA3 Q6NL41 A0A154PMZ9 A0A0L7RCF6 A0A2R7W199 A0A182IM49 A0A1S4JX30 E1ZV04 B4MC78 A0A0J7KH91 A0A182FMD6 A0A084VV02 E0W3L9 A0A1Q3FRP6 A0A2H8TJK6 A0A1J1HRG4 A0A2S2R3J0 A0A0K8ULM7 A0A0A1WQY2 A0A2S2NKQ7 A0A1A9WYM2 A0A1I8NID2 A0A0P5NBE9 A0A0P5NUT5 A0A0P4YKL2 A0A162T655 A0A182NAG7 A0A0P4ZAN5 A0A0P4XXB4 A0A0N8DEH6 A0A0P5YEQ8 A0A0P5LRD3 A0A0P5VS04 A0A0K8T4S3 A0A146LVX5 A0A0A9YIN9 A0A0P5ZI78
A0A194QBR9 A0A1W4WRJ1 A0A158NM21 A0A195CX99 A0A087ZQY3 F4WHS9 A0A0R3NMZ0 A0A067RHI5 A0A195BQV5 D6WAI6 A0A0M8ZQL2 B4N968 A0A026WEV7 A0A310SHN9 A0A151X0P3 A0A3B0KAZ0 A0A151JPZ4 E2B478 A0A0L0CME5 B5DWP1 A0A182U1N1 A0A182SQ16 A0A182WY99 A0A182VF87 A0A182HIH2 A0A1I8NMM5 A0A151JVU1 Q7QET1 U4U3Y9 N6UAV2 A0A182Q0W9 A0A0R1EE59 A0A336M410 A0A232F927 A0A0P8XWI3 A0A182YR91 A0A1B0B2J7 A0A1A9VFU1 A0A1A9YT10 B4K4F8 A0A1B0FI81 A0A224XLW9 T1HA98 A0A182MDE8 A0A182RL51 A0A182PV64 A0A0Q5WCU9 A0A0R1EBS8 A0A182W439 A0A182KAD8 A0A0R1E825 A0A0S0WGX2 A0A1B6JA19 A0A1B6HCC0 W8C3Y7 A0A023F2V7 A0A1B6D576 A0A1B6EXA3 Q6NL41 A0A154PMZ9 A0A0L7RCF6 A0A2R7W199 A0A182IM49 A0A1S4JX30 E1ZV04 B4MC78 A0A0J7KH91 A0A182FMD6 A0A084VV02 E0W3L9 A0A1Q3FRP6 A0A2H8TJK6 A0A1J1HRG4 A0A2S2R3J0 A0A0K8ULM7 A0A0A1WQY2 A0A2S2NKQ7 A0A1A9WYM2 A0A1I8NID2 A0A0P5NBE9 A0A0P5NUT5 A0A0P4YKL2 A0A162T655 A0A182NAG7 A0A0P4ZAN5 A0A0P4XXB4 A0A0N8DEH6 A0A0P5YEQ8 A0A0P5LRD3 A0A0P5VS04 A0A0K8T4S3 A0A146LVX5 A0A0A9YIN9 A0A0P5ZI78
Ontologies
GO
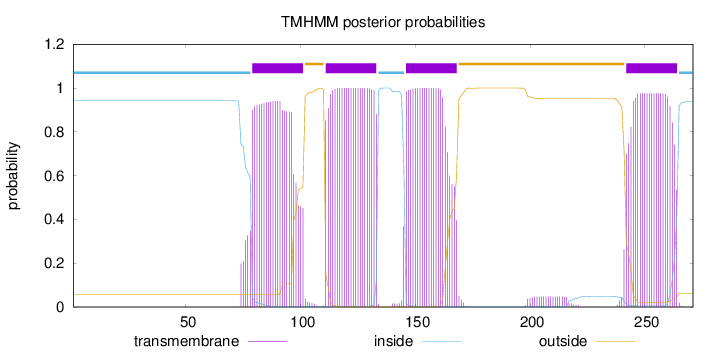
Topology
Length:
271
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.4322200000001
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.94140
inside
1 - 78
TMhelix
79 - 101
outside
102 - 110
TMhelix
111 - 133
inside
134 - 145
TMhelix
146 - 168
outside
169 - 241
TMhelix
242 - 264
inside
265 - 271
Population Genetic Test Statistics
Pi
197.712747
Theta
182.528193
Tajima's D
0.451604
CLR
1.570904
CSRT
0.502124893755312
Interpretation
Uncertain
