Gene
KWMTBOMO05216 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002430
Annotation
hypothetical_protein_KGM_03534_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.958
Sequence
CDS
ATGGGGTTAGATTTCTTAGAGCATGGGGCGGACTTTGAAGCCGCCATTTCGGCTACGGGTTTCGGTCGCTTCCACTTTTGCCTGCTTGCGGTCACGGGGCTGATCTACGCTAACACCGCCATTGGAATCACAATAGTGTCGTTCGTGCTACCGTCGGCGACATGTGATTTCCGCATGACTTCTGCGGACAAGGGCTGGCTTACAGCCGCGCCTATGTTGGGTATGGTCATTGGTTCATATTTCTGGGGTTGCTTGGCTGATACAAAGGGCCGTAAGGTTGTATTAGTATCAACCCTACTACTTGACGGCGTGGTCGGTATCATCTCATCGTTCGTCCAGATCTTGCCCATATTCATGGCTTGCAGATTCGTCAATGGTTTTGCCGTTGCCGGTGCTATGGGTATTTGCTTCCCGTACCTGGGAGAGTTCCAACAGACCAAATACAGAGAGAAAATCCTGTGCTGGATGGAAATGTTCTGGACTCTCGGTGTCATCTTGCTTCCCTTGATCGCCTGGGGCATCATTCCAATCCAAGGCATCCGCGTTGAGAGCGGTGCATTCTCCTACGACTCGTGGAACTGGTTCGTTGCTGCTTGTGGTGTTCCTTCCCTTCTCCTCGGTTTCTGGCTCTTCTCGTTCCCCGAGAGTCCCAAATTCATGATGGAGTGCGGAGACTACGACGATGCCCTGAAATGCCTGAAGAGCGTCTACAGACAAAACACCGGTGACGATCCGGATAATTATCCGATCAAGAGTCTAAAAGAGAAAGTGCGAACTGTGTCCGTTGCATCGCAAAGCTCCCAGAAATCCGTCAGAAGCCTTAGCATGCGCAAGCGGAAGGATCTGAAGCGTTTATTCACTGAGATTTGGACCCAGACCAAGGCTCTCTGCAGACCACCATACCTCAAATACACTATCTTGACTTGCCTGATTCAATTCGGCCTGACCACAAGTTACTACACCTTGATGATCTGGTTCCCCGAGCTGTTCAACCGTTACGAAGAGTTCGAACATCGTTTCCCCAACACATCAGCTTCGGTTTGCGACGTGTCCAGCATTGTCGTCGTTGACGAAGCATCGTCTGATCCCTTCGACTGCGAGAAGGTCATCGACCAGAGCGTCTACATGCACACCCTGATTGTCGGTCTCGCCTGCATTCCCACCTCGCTCTGGCTACCCCTCTGCGTTCATAAGCTTGGTGCCAAATTTTTCTTGATTTTCAGTTTGAGTTTCGCCGGAGTGGTGACAGTCGGTTTGTACTACGTACAGAACTCCATCCAGAATTTGGTACTGTCCTGTATCTTTGAAGCCCTCACCTCGCTCGCCATCAGTCTCGTGTTCTGCGTGCTCGTCGACCTGTTCCCTACAAATCTCAGGGTAATGGCTGCCGCGTTATCGCTGACTGCTGGTCGAGGTGGAGGTCTGATAGGAAACCTTTCGTTCGGTTACCTTATCGACTTGGCTTGTGTCGTACCAATTGTTGTTTTCTCAGCATTTTTATTCATTGCTGCTATACTCTGCTTCTTCTTGCCAAAGACGGGACAGGAAGCCTTGGACTAA
Protein
MGLDFLEHGADFEAAISATGFGRFHFCLLAVTGLIYANTAIGITIVSFVLPSATCDFRMTSADKGWLTAAPMLGMVIGSYFWGCLADTKGRKVVLVSTLLLDGVVGIISSFVQILPIFMACRFVNGFAVAGAMGICFPYLGEFQQTKYREKILCWMEMFWTLGVILLPLIAWGIIPIQGIRVESGAFSYDSWNWFVAACGVPSLLLGFWLFSFPESPKFMMECGDYDDALKCLKSVYRQNTGDDPDNYPIKSLKEKVRTVSVASQSSQKSVRSLSMRKRKDLKRLFTEIWTQTKALCRPPYLKYTILTCLIQFGLTTSYYTLMIWFPELFNRYEEFEHRFPNTSASVCDVSSIVVVDEASSDPFDCEKVIDQSVYMHTLIVGLACIPTSLWLPLCVHKLGAKFFLIFSLSFAGVVTVGLYYVQNSIQNLVLSCIFEALTSLAISLVFCVLVDLFPTNLRVMAAALSLTAGRGGGLIGNLSFGYLIDLACVVPIVVFSAFLFIAAILCFFLPKTGQEALD
Summary
Uniprot
H9IYU5
A0A212ESJ6
A0A2H1VMI9
A0A194RSD6
A0A194QBE5
A0A2A4J710
+ More
S4PYD7 D6WAI5 K7IYL5 A0A2A4J704 A0A154PLN8 A0A1B0CZJ6 A0A0C9R7R4 A0A336MF81 E2B477 A0A0L7RC69 A0A1Y1KR70 A0A026WFC5 A0A1Y1KNN4 E9J4Z0 A0A0M8ZS97 A0A158NM89 A0A232FF43 A0A151JVX9 A0A1S4FVL6 A0A084VV04 A0A1J1HTA4 A0A151JPY9 A0A182YQT2 A0A195CWY6 A0A0L0BTJ0 A0A1Q3FLS2 A0A1B6CG08 B0WWU3 Q16N29 A0A1A9WYM5 E1ZV02 A0A182FMD5 A0A182NAG2 A0A182J0Q5 N6U544 A0A151X0M7 A0A3B0K2C3 A0A182Q543 A0A182KCL9 A0A182VH93 F4WHS8 A0A2A3EJA3 A0A182HII0 A0A182PU98 Q7QES5 A0A1W4X2H9 A0A1W4WRJ2 A0A1W4X2A3 A0A195BRX6 A0A1Y1KTJ9 A0A0J7KHI2 A0A182W433 A0A182RVU8 A0A182WY91 A0A3B0KR71 J9JQX5 A0A2H8TTN8 A0A1B0FI37 A0A182S8V6 A0A182TDY5 A0A3B0K296 A0A1A9ZZ58 W5JU84 A0A336M5U0 A0A3L8DJU1 A0A2S2QJC6 A0A182MBH2 A0A2R7W2U3 T1HJP6 A0A146KZ19 A0A1I8PCK4 A0A087ZQS9 J9JTZ4 A0A139WMH7 A0A139WMB8 A0A2H8TEZ7 K7IYL4 A0A232FA58 A0A2S2NHU2 A0A182QYJ9 A0A182NC72 A0A3S2PIS8 J9K5L1 A0A182YKK5 A0A2H8TSI0 H9IW67 A0A182J0P8 A0A2M3ZF42
S4PYD7 D6WAI5 K7IYL5 A0A2A4J704 A0A154PLN8 A0A1B0CZJ6 A0A0C9R7R4 A0A336MF81 E2B477 A0A0L7RC69 A0A1Y1KR70 A0A026WFC5 A0A1Y1KNN4 E9J4Z0 A0A0M8ZS97 A0A158NM89 A0A232FF43 A0A151JVX9 A0A1S4FVL6 A0A084VV04 A0A1J1HTA4 A0A151JPY9 A0A182YQT2 A0A195CWY6 A0A0L0BTJ0 A0A1Q3FLS2 A0A1B6CG08 B0WWU3 Q16N29 A0A1A9WYM5 E1ZV02 A0A182FMD5 A0A182NAG2 A0A182J0Q5 N6U544 A0A151X0M7 A0A3B0K2C3 A0A182Q543 A0A182KCL9 A0A182VH93 F4WHS8 A0A2A3EJA3 A0A182HII0 A0A182PU98 Q7QES5 A0A1W4X2H9 A0A1W4WRJ2 A0A1W4X2A3 A0A195BRX6 A0A1Y1KTJ9 A0A0J7KHI2 A0A182W433 A0A182RVU8 A0A182WY91 A0A3B0KR71 J9JQX5 A0A2H8TTN8 A0A1B0FI37 A0A182S8V6 A0A182TDY5 A0A3B0K296 A0A1A9ZZ58 W5JU84 A0A336M5U0 A0A3L8DJU1 A0A2S2QJC6 A0A182MBH2 A0A2R7W2U3 T1HJP6 A0A146KZ19 A0A1I8PCK4 A0A087ZQS9 J9JTZ4 A0A139WMH7 A0A139WMB8 A0A2H8TEZ7 K7IYL4 A0A232FA58 A0A2S2NHU2 A0A182QYJ9 A0A182NC72 A0A3S2PIS8 J9K5L1 A0A182YKK5 A0A2H8TSI0 H9IW67 A0A182J0P8 A0A2M3ZF42
Pubmed
EMBL
BABH01010760
AGBW02012778
OWR44449.1
ODYU01003368
SOQ42041.1
KQ459984
+ More
KPJ18981.1 KQ459220 KPJ02863.1 NWSH01002966 PCG67203.1 GAIX01003468 JAA89092.1 KQ971312 EEZ97996.1 PCG67202.1 KQ434960 KZC12677.1 AJVK01009607 GBYB01004070 JAG73837.1 UFQT01000487 SSX24708.1 GL445515 EFN89494.1 KQ414616 KOC68423.1 GEZM01080373 GEZM01080372 JAV62185.1 KK107238 EZA54618.1 GEZM01080370 JAV62188.1 GL768121 EFZ12128.1 KQ435911 KOX68926.1 ADTU01020150 NNAY01000295 OXU29386.1 KQ981688 KYN37931.1 ATLV01017139 KE525157 KFB41798.1 CVRI01000020 CRK90612.1 KQ978684 KYN29305.1 KQ977171 KYN05178.1 JRES01001365 KNC23336.1 GFDL01006577 JAV28468.1 GEDC01025023 JAS12275.1 DS232152 EDS36204.1 CH477841 EAT35757.1 GL434335 EFN75000.1 APGK01042207 KB741002 ENN75761.1 KQ982610 KYQ53909.1 OUUW01000014 SPP88384.1 AXCN02002022 GL888167 EGI66285.1 KZ288226 PBC31873.1 APCN01003972 AAAB01008846 EAA06762.5 KQ976421 KYM89117.1 GEZM01080371 JAV62187.1 LBMM01007490 KMQ89702.1 SPP88386.1 ABLF02017432 ABLF02017434 ABLF02032144 GFXV01005504 MBW17309.1 CCAG010001331 SPP88387.1 ADMH02000432 ETN66510.1 UFQS01000488 UFQT01000488 SSX04347.1 SSX24711.1 QOIP01000007 RLU20667.1 GGMS01008652 MBY77855.1 AXCM01000645 AXCM01000646 KK854234 PTY13511.1 ACPB03019799 GDHC01017015 JAQ01614.1 ABLF02040460 KYB29083.1 KYB29082.1 GFXV01000868 MBW12673.1 NNAY01000620 OXU27360.1 GGMR01004128 MBY16747.1 AXCN02001759 RSAL01000020 RVE52607.1 ABLF02018816 GFXV01000848 GFXV01001356 GFXV01005006 GFXV01008039 MBW12653.1 MBW13161.1 MBW16811.1 MBW19844.1 BABH01005928 BABH01005929 BABH01005930 GGFM01006372 MBW27123.1
KPJ18981.1 KQ459220 KPJ02863.1 NWSH01002966 PCG67203.1 GAIX01003468 JAA89092.1 KQ971312 EEZ97996.1 PCG67202.1 KQ434960 KZC12677.1 AJVK01009607 GBYB01004070 JAG73837.1 UFQT01000487 SSX24708.1 GL445515 EFN89494.1 KQ414616 KOC68423.1 GEZM01080373 GEZM01080372 JAV62185.1 KK107238 EZA54618.1 GEZM01080370 JAV62188.1 GL768121 EFZ12128.1 KQ435911 KOX68926.1 ADTU01020150 NNAY01000295 OXU29386.1 KQ981688 KYN37931.1 ATLV01017139 KE525157 KFB41798.1 CVRI01000020 CRK90612.1 KQ978684 KYN29305.1 KQ977171 KYN05178.1 JRES01001365 KNC23336.1 GFDL01006577 JAV28468.1 GEDC01025023 JAS12275.1 DS232152 EDS36204.1 CH477841 EAT35757.1 GL434335 EFN75000.1 APGK01042207 KB741002 ENN75761.1 KQ982610 KYQ53909.1 OUUW01000014 SPP88384.1 AXCN02002022 GL888167 EGI66285.1 KZ288226 PBC31873.1 APCN01003972 AAAB01008846 EAA06762.5 KQ976421 KYM89117.1 GEZM01080371 JAV62187.1 LBMM01007490 KMQ89702.1 SPP88386.1 ABLF02017432 ABLF02017434 ABLF02032144 GFXV01005504 MBW17309.1 CCAG010001331 SPP88387.1 ADMH02000432 ETN66510.1 UFQS01000488 UFQT01000488 SSX04347.1 SSX24711.1 QOIP01000007 RLU20667.1 GGMS01008652 MBY77855.1 AXCM01000645 AXCM01000646 KK854234 PTY13511.1 ACPB03019799 GDHC01017015 JAQ01614.1 ABLF02040460 KYB29083.1 KYB29082.1 GFXV01000868 MBW12673.1 NNAY01000620 OXU27360.1 GGMR01004128 MBY16747.1 AXCN02001759 RSAL01000020 RVE52607.1 ABLF02018816 GFXV01000848 GFXV01001356 GFXV01005006 GFXV01008039 MBW12653.1 MBW13161.1 MBW16811.1 MBW19844.1 BABH01005928 BABH01005929 BABH01005930 GGFM01006372 MBW27123.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000007266
+ More
UP000002358 UP000076502 UP000092462 UP000008237 UP000053825 UP000053097 UP000053105 UP000005205 UP000215335 UP000078541 UP000030765 UP000183832 UP000078492 UP000076408 UP000078542 UP000037069 UP000002320 UP000008820 UP000091820 UP000000311 UP000069272 UP000075884 UP000075880 UP000019118 UP000075809 UP000268350 UP000075886 UP000075881 UP000075903 UP000007755 UP000242457 UP000075840 UP000075885 UP000007062 UP000192223 UP000078540 UP000036403 UP000075920 UP000075900 UP000076407 UP000007819 UP000092444 UP000075901 UP000075902 UP000092445 UP000000673 UP000279307 UP000075883 UP000015103 UP000095300 UP000005203 UP000283053
UP000002358 UP000076502 UP000092462 UP000008237 UP000053825 UP000053097 UP000053105 UP000005205 UP000215335 UP000078541 UP000030765 UP000183832 UP000078492 UP000076408 UP000078542 UP000037069 UP000002320 UP000008820 UP000091820 UP000000311 UP000069272 UP000075884 UP000075880 UP000019118 UP000075809 UP000268350 UP000075886 UP000075881 UP000075903 UP000007755 UP000242457 UP000075840 UP000075885 UP000007062 UP000192223 UP000078540 UP000036403 UP000075920 UP000075900 UP000076407 UP000007819 UP000092444 UP000075901 UP000075902 UP000092445 UP000000673 UP000279307 UP000075883 UP000015103 UP000095300 UP000005203 UP000283053
Interpro
CDD
ProteinModelPortal
H9IYU5
A0A212ESJ6
A0A2H1VMI9
A0A194RSD6
A0A194QBE5
A0A2A4J710
+ More
S4PYD7 D6WAI5 K7IYL5 A0A2A4J704 A0A154PLN8 A0A1B0CZJ6 A0A0C9R7R4 A0A336MF81 E2B477 A0A0L7RC69 A0A1Y1KR70 A0A026WFC5 A0A1Y1KNN4 E9J4Z0 A0A0M8ZS97 A0A158NM89 A0A232FF43 A0A151JVX9 A0A1S4FVL6 A0A084VV04 A0A1J1HTA4 A0A151JPY9 A0A182YQT2 A0A195CWY6 A0A0L0BTJ0 A0A1Q3FLS2 A0A1B6CG08 B0WWU3 Q16N29 A0A1A9WYM5 E1ZV02 A0A182FMD5 A0A182NAG2 A0A182J0Q5 N6U544 A0A151X0M7 A0A3B0K2C3 A0A182Q543 A0A182KCL9 A0A182VH93 F4WHS8 A0A2A3EJA3 A0A182HII0 A0A182PU98 Q7QES5 A0A1W4X2H9 A0A1W4WRJ2 A0A1W4X2A3 A0A195BRX6 A0A1Y1KTJ9 A0A0J7KHI2 A0A182W433 A0A182RVU8 A0A182WY91 A0A3B0KR71 J9JQX5 A0A2H8TTN8 A0A1B0FI37 A0A182S8V6 A0A182TDY5 A0A3B0K296 A0A1A9ZZ58 W5JU84 A0A336M5U0 A0A3L8DJU1 A0A2S2QJC6 A0A182MBH2 A0A2R7W2U3 T1HJP6 A0A146KZ19 A0A1I8PCK4 A0A087ZQS9 J9JTZ4 A0A139WMH7 A0A139WMB8 A0A2H8TEZ7 K7IYL4 A0A232FA58 A0A2S2NHU2 A0A182QYJ9 A0A182NC72 A0A3S2PIS8 J9K5L1 A0A182YKK5 A0A2H8TSI0 H9IW67 A0A182J0P8 A0A2M3ZF42
S4PYD7 D6WAI5 K7IYL5 A0A2A4J704 A0A154PLN8 A0A1B0CZJ6 A0A0C9R7R4 A0A336MF81 E2B477 A0A0L7RC69 A0A1Y1KR70 A0A026WFC5 A0A1Y1KNN4 E9J4Z0 A0A0M8ZS97 A0A158NM89 A0A232FF43 A0A151JVX9 A0A1S4FVL6 A0A084VV04 A0A1J1HTA4 A0A151JPY9 A0A182YQT2 A0A195CWY6 A0A0L0BTJ0 A0A1Q3FLS2 A0A1B6CG08 B0WWU3 Q16N29 A0A1A9WYM5 E1ZV02 A0A182FMD5 A0A182NAG2 A0A182J0Q5 N6U544 A0A151X0M7 A0A3B0K2C3 A0A182Q543 A0A182KCL9 A0A182VH93 F4WHS8 A0A2A3EJA3 A0A182HII0 A0A182PU98 Q7QES5 A0A1W4X2H9 A0A1W4WRJ2 A0A1W4X2A3 A0A195BRX6 A0A1Y1KTJ9 A0A0J7KHI2 A0A182W433 A0A182RVU8 A0A182WY91 A0A3B0KR71 J9JQX5 A0A2H8TTN8 A0A1B0FI37 A0A182S8V6 A0A182TDY5 A0A3B0K296 A0A1A9ZZ58 W5JU84 A0A336M5U0 A0A3L8DJU1 A0A2S2QJC6 A0A182MBH2 A0A2R7W2U3 T1HJP6 A0A146KZ19 A0A1I8PCK4 A0A087ZQS9 J9JTZ4 A0A139WMH7 A0A139WMB8 A0A2H8TEZ7 K7IYL4 A0A232FA58 A0A2S2NHU2 A0A182QYJ9 A0A182NC72 A0A3S2PIS8 J9K5L1 A0A182YKK5 A0A2H8TSI0 H9IW67 A0A182J0P8 A0A2M3ZF42
PDB
4LDS
E-value=6.85069e-12,
Score=172
Ontologies
GO
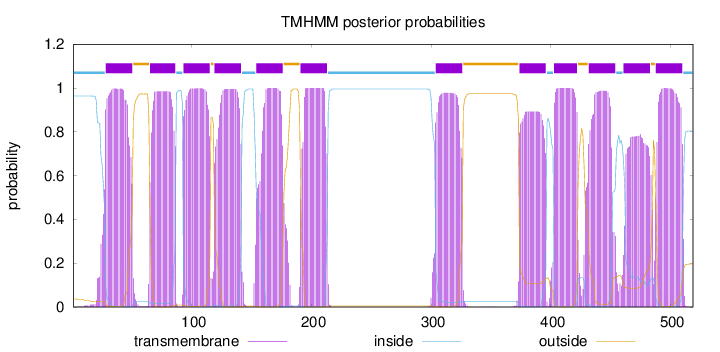
Topology
Length:
519
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
257.53385
Exp number, first 60 AAs:
23.69832
Total prob of N-in:
0.96359
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 64
TMhelix
65 - 86
inside
87 - 92
TMhelix
93 - 115
outside
116 - 118
TMhelix
119 - 141
inside
142 - 153
TMhelix
154 - 176
outside
177 - 190
TMhelix
191 - 213
inside
214 - 303
TMhelix
304 - 326
outside
327 - 373
TMhelix
374 - 396
inside
397 - 402
TMhelix
403 - 422
outside
423 - 431
TMhelix
432 - 454
inside
455 - 460
TMhelix
461 - 483
outside
484 - 487
TMhelix
488 - 510
inside
511 - 519
Population Genetic Test Statistics
Pi
242.221685
Theta
185.055502
Tajima's D
0.969181
CLR
0.080926
CSRT
0.653617319134043
Interpretation
Uncertain
