Gene
KWMTBOMO05215 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002470
Annotation
PREDICTED:_charged_multivesicular_body_protein_5_[Amyelois_transitella]
Full name
Charged multivesicular body protein 5
Alternative Name
Vacuolar protein-sorting-associated protein 60
Location in the cell
Cytoplasmic Reliability : 2.145 Nuclear Reliability : 2.59
Sequence
CDS
ATGAACAGATTATTTGGAAGAGCGAAACCCAAAGAACCTGGCCCCAGCATTACGGATTGCATTAAAAACGTAGATGGCCGAGCCGATAATATTGAACAGAAAGTCCAGAAACTGGACACAGAACTTAGAAAATACAAGGATCAGATGAGCAAAATGAGGGAGGGTCCTGCTAAAAATTCAGTCAAGCAAAAAGCAATGAGAGTATTGAAACAGAAGAAAATGTATGAACAACAACTTGATAATCTCCGTGCCCAGTCTTTCAACATGGAACAAGCAAATTATGCAACGCAGACATTGAAGGACACTCATACCACAATATCTGCAATGAAGGATGGTGTAACACAAATGAAAAAAGAATTTAAGAAGATTAATATTGATTCCATTGATGATGTAAATGATGAGTTAGCTGATATGATGGAACAAGCTGATGAAGTTCAAGAAGCCCTCGGCAGGCAATATGGTATGCCAGAGATTGATGATGATGAGCTTGCTGCAGAACTTGATGCTCTTGGTGATGAAATTGCTTTAGATGATGATGCTTCTTACCTTGACGATGTTGTGAAGGCTCCAGCAGCTCCAGATAGGGAACCTGGAGCCGACAGCATCCGGAACAAAGATGGTGTTCCCGTGGACGAATTTGGTCTGCCTCAGGTTCCAAGTACACCACAAACACACTGA
Protein
MNRLFGRAKPKEPGPSITDCIKNVDGRADNIEQKVQKLDTELRKYKDQMSKMREGPAKNSVKQKAMRVLKQKKMYEQQLDNLRAQSFNMEQANYATQTLKDTHTTISAMKDGVTQMKKEFKKINIDSIDDVNDELADMMEQADEVQEALGRQYGMPEIDDDELAAELDALGDEIALDDDASYLDDVVKAPAAPDREPGADSIRNKDGVPVDEFGLPQVPSTPQTH
Summary
Description
Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and are delivered to lysosomes enabling degradation of membrane proteins (By similarity). Specifically down-regulates Notch signaling activity in the germarium, probably by facilitating Notch endocytosis (PubMed:24762813).
Subunit
Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III).
Similarity
Belongs to the SNF7 family.
Keywords
Coiled coil
Complete proteome
Endosome
Membrane
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Charged multivesicular body protein 5
Uniprot
H9IYY5
A0A1V0JHU4
A0A2H1VMI4
A0A1E1W8T1
A0A194RNS7
A0A194QBM2
+ More
A0A212ESJ1 A0A1B0DMH6 A0A023EK64 Q1HQE2 A0A154PNF8 A0A2A3EI90 A0A088ACS6 E2BKB7 A0A026WAV5 A0A182JFU8 A0A182UNG6 A0A182TID4 A0A182X491 A0A182LBA3 Q7Q9Q8 A0A182HP94 E2AVJ2 A0A182T031 A0A0K8TTJ4 A0A182N897 A0A182RTL9 A0A182YFM5 A0A182P6B0 A0A182WIJ5 A0A182MB39 A0A2M4AH73 W5JVB9 B0XB50 A0A232FDQ3 K7IY65 A0A084W2J3 A0A182Q7T7 B0X7X3 U5EX55 T1DNI7 A0A2M3ZHU1 A0A2M4BYZ9 D6WCX4 A0A1Q3FHL6 A0A182FCF1 A0A1A9WP18 A0A1B0AI28 D3TNV0 A0A0L0C2Q4 A0A067QKH8 A0A1L8DVU0 A0A1A9VYQ7 A0A034VSS6 A0A0L7RHP8 A0A1A9XC91 A0A1B0B596 A0A1W4VSK8 A0A0K8V4E2 A0A1B0CJF1 B4J3J1 A0A0A1XT40 A0A1J1ISZ1 B4PJL2 B3NHT2 B4QNX4 A0A310STT2 A0A336MXA6 M9PCV9 Q9VVI9 A0A0T6ATI7 W8AUJ9 V5GZL1 F4W952 A0A1B6CQK7 E9G1P0 A0A158P084 A0A2J7PP90 T1PDW4 A0A1B6F678 A0A1W4WD55 A0A162T5J7 A0A023EMS9 B4N4D3 A0A0P5W573 A0A0N8BPF9 A0A3B0KI13 B4LEZ3 B3M4S5 A0A0P5EB27 A0A0P5CHR5 A0A1B6LEJ6 A0A1L8EEH2 Q2M0U1 A0A0P4X6B3 B4KZJ4 A0A1Y1M490 A0A1I8Q6U6 A0A0P4WE76 A0A0P5HBT5
A0A212ESJ1 A0A1B0DMH6 A0A023EK64 Q1HQE2 A0A154PNF8 A0A2A3EI90 A0A088ACS6 E2BKB7 A0A026WAV5 A0A182JFU8 A0A182UNG6 A0A182TID4 A0A182X491 A0A182LBA3 Q7Q9Q8 A0A182HP94 E2AVJ2 A0A182T031 A0A0K8TTJ4 A0A182N897 A0A182RTL9 A0A182YFM5 A0A182P6B0 A0A182WIJ5 A0A182MB39 A0A2M4AH73 W5JVB9 B0XB50 A0A232FDQ3 K7IY65 A0A084W2J3 A0A182Q7T7 B0X7X3 U5EX55 T1DNI7 A0A2M3ZHU1 A0A2M4BYZ9 D6WCX4 A0A1Q3FHL6 A0A182FCF1 A0A1A9WP18 A0A1B0AI28 D3TNV0 A0A0L0C2Q4 A0A067QKH8 A0A1L8DVU0 A0A1A9VYQ7 A0A034VSS6 A0A0L7RHP8 A0A1A9XC91 A0A1B0B596 A0A1W4VSK8 A0A0K8V4E2 A0A1B0CJF1 B4J3J1 A0A0A1XT40 A0A1J1ISZ1 B4PJL2 B3NHT2 B4QNX4 A0A310STT2 A0A336MXA6 M9PCV9 Q9VVI9 A0A0T6ATI7 W8AUJ9 V5GZL1 F4W952 A0A1B6CQK7 E9G1P0 A0A158P084 A0A2J7PP90 T1PDW4 A0A1B6F678 A0A1W4WD55 A0A162T5J7 A0A023EMS9 B4N4D3 A0A0P5W573 A0A0N8BPF9 A0A3B0KI13 B4LEZ3 B3M4S5 A0A0P5EB27 A0A0P5CHR5 A0A1B6LEJ6 A0A1L8EEH2 Q2M0U1 A0A0P4X6B3 B4KZJ4 A0A1Y1M490 A0A1I8Q6U6 A0A0P4WE76 A0A0P5HBT5
Pubmed
19121390
26354079
22118469
24945155
17204158
17510324
+ More
20798317 24508170 30249741 20966253 12364791 14747013 17210077 26369729 25244985 20920257 23761445 28648823 20075255 24438588 18362917 19820115 20353571 26108605 24845553 25348373 17994087 25830018 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17372656 18327897 24762813 24495485 21719571 21292972 21347285 25315136 15632085 28004739
20798317 24508170 30249741 20966253 12364791 14747013 17210077 26369729 25244985 20920257 23761445 28648823 20075255 24438588 18362917 19820115 20353571 26108605 24845553 25348373 17994087 25830018 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17372656 18327897 24762813 24495485 21719571 21292972 21347285 25315136 15632085 28004739
EMBL
BABH01010760
KY694523
ARD08864.1
ODYU01003368
SOQ42040.1
GDQN01007695
+ More
JAT83359.1 KQ459984 KPJ18980.1 KQ459220 KPJ02862.1 AGBW02012778 OWR44450.1 AJVK01001342 GAPW01004085 JAC09513.1 DQ440502 CH477862 CH477360 ABF18535.1 EAT35527.1 EAT42670.1 KQ434999 KZC13372.1 KZ288234 PBC31437.1 GL448783 EFN83841.1 KK107293 QOIP01000005 EZA53232.1 RLU22570.1 AAAB01008900 EAA09450.2 APCN01002931 GL443122 EFN62543.1 GDAI01000343 JAI17260.1 AXCM01004441 GGFK01006766 MBW40087.1 ADMH02000391 ETN66744.1 DS232614 EDS44103.1 NNAY01000399 OXU28650.1 ATLV01019621 KE525275 KFB44437.1 AXCN02000962 DS232468 EDS42192.1 GANO01001120 JAB58751.1 GAMD01002835 JAA98755.1 GGFM01007330 MBW28081.1 GGFJ01009112 MBW58253.1 KQ971311 EEZ98827.1 GFDL01007989 JAV27056.1 CCAG010009787 EZ423102 ADD19378.1 JRES01001077 KNC25699.1 KK853234 KDR09590.1 GFDF01003536 JAV10548.1 GAKP01013438 JAC45514.1 KQ414590 KOC70390.1 JXJN01008650 GDHF01033214 GDHF01032892 GDHF01018532 GDHF01018174 GDHF01011465 JAI19100.1 JAI19422.1 JAI33782.1 JAI34140.1 JAI40849.1 AJWK01014542 AJWK01014543 CH916366 EDV96193.1 GBXI01011467 GBXI01000187 JAD02825.1 JAD14105.1 CVRI01000057 CRL02222.1 CM000159 EDW94700.1 CH954178 EDV51947.1 CM000363 CM002912 EDX10876.1 KMZ00275.1 KQ760478 OAD60230.1 UFQS01003338 UFQS01004223 UFQT01003338 UFQT01004223 SSX15520.1 SSX16260.1 SSX34886.1 AE014296 AGB94687.1 AY071206 LJIG01022880 KRT78253.1 GAMC01014065 JAB92490.1 GALX01002543 JAB65923.1 GL888002 EGI69234.1 GEDC01021640 GEDC01016924 GEDC01004251 JAS15658.1 JAS20374.1 JAS33047.1 GL732529 EFX86777.1 ADTU01000980 NEVH01023278 PNF18162.1 KA646330 AFP60959.1 GECZ01024000 JAS45769.1 LRGB01000007 KZS21926.1 GAPW01003949 JAC09649.1 CH964095 EDW79007.1 GDIP01091068 JAM12647.1 GDIQ01157787 JAK93938.1 OUUW01000009 SPP84751.1 CH940647 EDW69162.1 CH902618 EDV40499.1 GDIP01144111 JAJ79291.1 GDIP01186053 JAJ37349.1 GEBQ01018053 JAT21924.1 GFDG01001800 JAV16999.1 CH379069 EAL30835.1 GDIP01246363 JAI77038.1 CH933809 EDW17921.1 GEZM01041009 JAV80654.1 GDRN01042492 JAI67043.1 GDIQ01230109 JAK21616.1
JAT83359.1 KQ459984 KPJ18980.1 KQ459220 KPJ02862.1 AGBW02012778 OWR44450.1 AJVK01001342 GAPW01004085 JAC09513.1 DQ440502 CH477862 CH477360 ABF18535.1 EAT35527.1 EAT42670.1 KQ434999 KZC13372.1 KZ288234 PBC31437.1 GL448783 EFN83841.1 KK107293 QOIP01000005 EZA53232.1 RLU22570.1 AAAB01008900 EAA09450.2 APCN01002931 GL443122 EFN62543.1 GDAI01000343 JAI17260.1 AXCM01004441 GGFK01006766 MBW40087.1 ADMH02000391 ETN66744.1 DS232614 EDS44103.1 NNAY01000399 OXU28650.1 ATLV01019621 KE525275 KFB44437.1 AXCN02000962 DS232468 EDS42192.1 GANO01001120 JAB58751.1 GAMD01002835 JAA98755.1 GGFM01007330 MBW28081.1 GGFJ01009112 MBW58253.1 KQ971311 EEZ98827.1 GFDL01007989 JAV27056.1 CCAG010009787 EZ423102 ADD19378.1 JRES01001077 KNC25699.1 KK853234 KDR09590.1 GFDF01003536 JAV10548.1 GAKP01013438 JAC45514.1 KQ414590 KOC70390.1 JXJN01008650 GDHF01033214 GDHF01032892 GDHF01018532 GDHF01018174 GDHF01011465 JAI19100.1 JAI19422.1 JAI33782.1 JAI34140.1 JAI40849.1 AJWK01014542 AJWK01014543 CH916366 EDV96193.1 GBXI01011467 GBXI01000187 JAD02825.1 JAD14105.1 CVRI01000057 CRL02222.1 CM000159 EDW94700.1 CH954178 EDV51947.1 CM000363 CM002912 EDX10876.1 KMZ00275.1 KQ760478 OAD60230.1 UFQS01003338 UFQS01004223 UFQT01003338 UFQT01004223 SSX15520.1 SSX16260.1 SSX34886.1 AE014296 AGB94687.1 AY071206 LJIG01022880 KRT78253.1 GAMC01014065 JAB92490.1 GALX01002543 JAB65923.1 GL888002 EGI69234.1 GEDC01021640 GEDC01016924 GEDC01004251 JAS15658.1 JAS20374.1 JAS33047.1 GL732529 EFX86777.1 ADTU01000980 NEVH01023278 PNF18162.1 KA646330 AFP60959.1 GECZ01024000 JAS45769.1 LRGB01000007 KZS21926.1 GAPW01003949 JAC09649.1 CH964095 EDW79007.1 GDIP01091068 JAM12647.1 GDIQ01157787 JAK93938.1 OUUW01000009 SPP84751.1 CH940647 EDW69162.1 CH902618 EDV40499.1 GDIP01144111 JAJ79291.1 GDIP01186053 JAJ37349.1 GEBQ01018053 JAT21924.1 GFDG01001800 JAV16999.1 CH379069 EAL30835.1 GDIP01246363 JAI77038.1 CH933809 EDW17921.1 GEZM01041009 JAV80654.1 GDRN01042492 JAI67043.1 GDIQ01230109 JAK21616.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000092462
UP000008820
+ More
UP000076502 UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000075880 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000311 UP000075901 UP000075884 UP000075900 UP000076408 UP000075885 UP000075920 UP000075883 UP000000673 UP000002320 UP000215335 UP000002358 UP000030765 UP000075886 UP000007266 UP000069272 UP000091820 UP000092445 UP000092444 UP000037069 UP000027135 UP000078200 UP000053825 UP000092443 UP000092460 UP000192221 UP000092461 UP000001070 UP000183832 UP000002282 UP000008711 UP000000304 UP000000803 UP000007755 UP000000305 UP000005205 UP000235965 UP000095301 UP000192223 UP000076858 UP000007798 UP000268350 UP000008792 UP000007801 UP000001819 UP000009192 UP000095300
UP000076502 UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000075880 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000311 UP000075901 UP000075884 UP000075900 UP000076408 UP000075885 UP000075920 UP000075883 UP000000673 UP000002320 UP000215335 UP000002358 UP000030765 UP000075886 UP000007266 UP000069272 UP000091820 UP000092445 UP000092444 UP000037069 UP000027135 UP000078200 UP000053825 UP000092443 UP000092460 UP000192221 UP000092461 UP000001070 UP000183832 UP000002282 UP000008711 UP000000304 UP000000803 UP000007755 UP000000305 UP000005205 UP000235965 UP000095301 UP000192223 UP000076858 UP000007798 UP000268350 UP000008792 UP000007801 UP000001819 UP000009192 UP000095300
Pfam
PF03357 Snf7
Interpro
IPR005024
Snf7_fam
ProteinModelPortal
H9IYY5
A0A1V0JHU4
A0A2H1VMI4
A0A1E1W8T1
A0A194RNS7
A0A194QBM2
+ More
A0A212ESJ1 A0A1B0DMH6 A0A023EK64 Q1HQE2 A0A154PNF8 A0A2A3EI90 A0A088ACS6 E2BKB7 A0A026WAV5 A0A182JFU8 A0A182UNG6 A0A182TID4 A0A182X491 A0A182LBA3 Q7Q9Q8 A0A182HP94 E2AVJ2 A0A182T031 A0A0K8TTJ4 A0A182N897 A0A182RTL9 A0A182YFM5 A0A182P6B0 A0A182WIJ5 A0A182MB39 A0A2M4AH73 W5JVB9 B0XB50 A0A232FDQ3 K7IY65 A0A084W2J3 A0A182Q7T7 B0X7X3 U5EX55 T1DNI7 A0A2M3ZHU1 A0A2M4BYZ9 D6WCX4 A0A1Q3FHL6 A0A182FCF1 A0A1A9WP18 A0A1B0AI28 D3TNV0 A0A0L0C2Q4 A0A067QKH8 A0A1L8DVU0 A0A1A9VYQ7 A0A034VSS6 A0A0L7RHP8 A0A1A9XC91 A0A1B0B596 A0A1W4VSK8 A0A0K8V4E2 A0A1B0CJF1 B4J3J1 A0A0A1XT40 A0A1J1ISZ1 B4PJL2 B3NHT2 B4QNX4 A0A310STT2 A0A336MXA6 M9PCV9 Q9VVI9 A0A0T6ATI7 W8AUJ9 V5GZL1 F4W952 A0A1B6CQK7 E9G1P0 A0A158P084 A0A2J7PP90 T1PDW4 A0A1B6F678 A0A1W4WD55 A0A162T5J7 A0A023EMS9 B4N4D3 A0A0P5W573 A0A0N8BPF9 A0A3B0KI13 B4LEZ3 B3M4S5 A0A0P5EB27 A0A0P5CHR5 A0A1B6LEJ6 A0A1L8EEH2 Q2M0U1 A0A0P4X6B3 B4KZJ4 A0A1Y1M490 A0A1I8Q6U6 A0A0P4WE76 A0A0P5HBT5
A0A212ESJ1 A0A1B0DMH6 A0A023EK64 Q1HQE2 A0A154PNF8 A0A2A3EI90 A0A088ACS6 E2BKB7 A0A026WAV5 A0A182JFU8 A0A182UNG6 A0A182TID4 A0A182X491 A0A182LBA3 Q7Q9Q8 A0A182HP94 E2AVJ2 A0A182T031 A0A0K8TTJ4 A0A182N897 A0A182RTL9 A0A182YFM5 A0A182P6B0 A0A182WIJ5 A0A182MB39 A0A2M4AH73 W5JVB9 B0XB50 A0A232FDQ3 K7IY65 A0A084W2J3 A0A182Q7T7 B0X7X3 U5EX55 T1DNI7 A0A2M3ZHU1 A0A2M4BYZ9 D6WCX4 A0A1Q3FHL6 A0A182FCF1 A0A1A9WP18 A0A1B0AI28 D3TNV0 A0A0L0C2Q4 A0A067QKH8 A0A1L8DVU0 A0A1A9VYQ7 A0A034VSS6 A0A0L7RHP8 A0A1A9XC91 A0A1B0B596 A0A1W4VSK8 A0A0K8V4E2 A0A1B0CJF1 B4J3J1 A0A0A1XT40 A0A1J1ISZ1 B4PJL2 B3NHT2 B4QNX4 A0A310STT2 A0A336MXA6 M9PCV9 Q9VVI9 A0A0T6ATI7 W8AUJ9 V5GZL1 F4W952 A0A1B6CQK7 E9G1P0 A0A158P084 A0A2J7PP90 T1PDW4 A0A1B6F678 A0A1W4WD55 A0A162T5J7 A0A023EMS9 B4N4D3 A0A0P5W573 A0A0N8BPF9 A0A3B0KI13 B4LEZ3 B3M4S5 A0A0P5EB27 A0A0P5CHR5 A0A1B6LEJ6 A0A1L8EEH2 Q2M0U1 A0A0P4X6B3 B4KZJ4 A0A1Y1M490 A0A1I8Q6U6 A0A0P4WE76 A0A0P5HBT5
PDB
5FD7
E-value=5.55654e-05,
Score=108
Ontologies
GO
Topology
Subcellular location
Endosome membrane
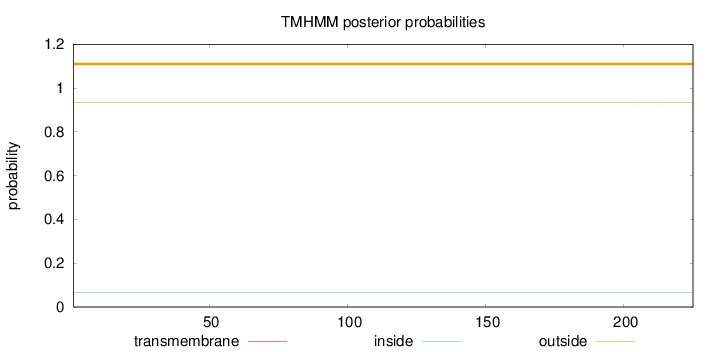
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06582
outside
1 - 225
Population Genetic Test Statistics
Pi
209.151101
Theta
144.903482
Tajima's D
1.70267
CLR
0.235313
CSRT
0.828758562071896
Interpretation
Uncertain
