Gene
KWMTBOMO05214
Pre Gene Modal
BGIBMGA002431
Annotation
PREDICTED:_thiamin_pyrophosphokinase_1_[Bombyx_mori]
Full name
Thiamine pyrophosphokinase
+ More
Thiamin pyrophosphokinase 1
Thiamin pyrophosphokinase 1
Alternative Name
Thiamine pyrophosphokinase 1
Location in the cell
Cytoplasmic Reliability : 2.706
Sequence
CDS
ATGCACAATAAACCTAGTTTAAGATATGCAGTTCTCATTTTAAATAGACCAATATCACAAGAAGAAGCATTTATGAAAAACTTTTGGGAAAAAGCCACATTAAGAATAACAGTAGATGGTGGAGCATTAAGATGGGACCAGTTTCTCACCAGACTACCTGAGGAAATTAGTAATAAAATGAAAATACCAGATTTGATTACTGGTGATTTTGATTCAATAACAGAAGAAACTCTCCAAAAATATAAAAAGAAAGGTTGTAAGGTAGTGCACACCCCAGATCAAGATTATACAGATTTTACAAAAGCACTGAAAGAGTTGAATAATCACTGTCAAGAACATACAATTGAGGTGGACAATGTAATAGCAATAGGACAGGCTTCAGGCAGACTAGACCAAATCTTAGGAAATATACAAACTTTATATCTAGCTGAAAAACAACATTTACTTCATCCTGAAACTAAACTCTACCTAATGTCCGGTGATTCATTATCTTGGCTATTGGAACCTGGCAATCATATTATAAGCATTCCCGAGGAAACAAGGAAGCATAACAGAGCTTGGTGTTCATTAGTGCCTATTGGAGAAACTTGCCACAGTGTCACAACAAGTGGATTGATGTGGAATTTAGACAATCAAAGCTTGAAATTTGGAGAACTAGTCAGCACGTCGAATGCATTCGATGGTTCAGACACAGTCAAGATCAAATGTAGCCACACAATTCTTTGGTCTATGAAGGTGCCCAGTTTGACAGGAGAATAA
Protein
MHNKPSLRYAVLILNRPISQEEAFMKNFWEKATLRITVDGGALRWDQFLTRLPEEISNKMKIPDLITGDFDSITEETLQKYKKKGCKVVHTPDQDYTDFTKALKELNNHCQEHTIEVDNVIAIGQASGRLDQILGNIQTLYLAEKQHLLHPETKLYLMSGDSLSWLLEPGNHIISIPEETRKHNRAWCSLVPIGETCHSVTTSGLMWNLDNQSLKFGELVSTSNAFDGSDTVKIKCSHTILWSMKVPSLTGE
Summary
Description
Catalyzes the phosphorylation of thiamine to thiamine pyrophosphate. Can also catalyze the phosphorylation of pyrithiamine to pyrithiamine pyrophosphate.
Catalytic Activity
ATP + thiamine = AMP + H(+) + thiamine diphosphate
Subunit
Homodimer.
Similarity
Belongs to the thiamine pyrophosphokinase family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Keywords
3D-structure
Alternative splicing
ATP-binding
Complete proteome
Kinase
Nucleotide-binding
Reference proteome
Transferase
Feature
chain Thiamin pyrophosphokinase 1
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A194QHE2
A0A2H1VMH9
A0A212ESN4
A0A3S2LCP0
A0A2A4J547
A0A194RNJ0
+ More
V5I9I0 A0A1B0CAN8 B0X7F9 A0A1Q3FJT3 A0A1Y1JTR1 A0A182FNN9 A0A2M4AHT2 W5JJV2 A0A2M4AH18 A0A2M4AGZ1 A0A2M4AHY5 A0A2M4BWQ1 T1E9W7 A0A067RJS6 A0A2M4BVU1 A0A2M4BWX1 T1GTI5 A0A2M4BXC7 J3JUK3 A0A131XFB5 A0A1Y9HFB6 A0A2M3ZC20 C1BVE9 D3PJQ4 A0A023FP32 A0A182JUP7 U5EJ18 Q175Q4 A0A0K8TMN4 A0A182MFE1 A0A224Z5K2 L7LZG3 A0A023GBB3 A0A131Z148 A0A023ELM5 G3MR72 A0A2R5L890 A0A2K8JX53 C1BTH3 A0A182UUV8 A0A0K8R8Y1 A0A1Y9IVZ1 A0A182WRQ4 A0A182U6J6 A0A1A9WJK7 A0A182HU24 A0A182LG03 D6W952 A0A0L0CS03 Q7QD56 A0A139WMT9 A0A182P6N6 K7J2E2 A0A2P8XMV1 A0A131XYR3 A0A310SEX6 A0A154PNU9 A0A1B0AAB9 A0A1A9UN97 A0A182XWD1 A0A1I8Q7U0 Q28HS1 A0A1B0G6V7 F6TP13 A0A084VZB1 A0A232ENS3 A0A293MYH5 A0A240PNY2 A0A182SMJ9 A0A151WZI5 F4W4E8 A0A1B6DCN3 G3HW81 A0A1W4X2H6 A0A0L7QWN9 A0A182QE81 A0A1L8FWN4 A0A0T6BC94 G3QVB4 W5QAX1 F7EK85 Q9R0M5 A0A1Z5L5K6 A0A0C9RUM1 E2BB74 A0A2U3WL60 A0A2K5VA75 E9JC14 A0A2I2ZCZ0 A0A286X7B8 I3N387 G1P1W6
V5I9I0 A0A1B0CAN8 B0X7F9 A0A1Q3FJT3 A0A1Y1JTR1 A0A182FNN9 A0A2M4AHT2 W5JJV2 A0A2M4AH18 A0A2M4AGZ1 A0A2M4AHY5 A0A2M4BWQ1 T1E9W7 A0A067RJS6 A0A2M4BVU1 A0A2M4BWX1 T1GTI5 A0A2M4BXC7 J3JUK3 A0A131XFB5 A0A1Y9HFB6 A0A2M3ZC20 C1BVE9 D3PJQ4 A0A023FP32 A0A182JUP7 U5EJ18 Q175Q4 A0A0K8TMN4 A0A182MFE1 A0A224Z5K2 L7LZG3 A0A023GBB3 A0A131Z148 A0A023ELM5 G3MR72 A0A2R5L890 A0A2K8JX53 C1BTH3 A0A182UUV8 A0A0K8R8Y1 A0A1Y9IVZ1 A0A182WRQ4 A0A182U6J6 A0A1A9WJK7 A0A182HU24 A0A182LG03 D6W952 A0A0L0CS03 Q7QD56 A0A139WMT9 A0A182P6N6 K7J2E2 A0A2P8XMV1 A0A131XYR3 A0A310SEX6 A0A154PNU9 A0A1B0AAB9 A0A1A9UN97 A0A182XWD1 A0A1I8Q7U0 Q28HS1 A0A1B0G6V7 F6TP13 A0A084VZB1 A0A232ENS3 A0A293MYH5 A0A240PNY2 A0A182SMJ9 A0A151WZI5 F4W4E8 A0A1B6DCN3 G3HW81 A0A1W4X2H6 A0A0L7QWN9 A0A182QE81 A0A1L8FWN4 A0A0T6BC94 G3QVB4 W5QAX1 F7EK85 Q9R0M5 A0A1Z5L5K6 A0A0C9RUM1 E2BB74 A0A2U3WL60 A0A2K5VA75 E9JC14 A0A2I2ZCZ0 A0A286X7B8 I3N387 G1P1W6
EC Number
2.7.6.2
Pubmed
26354079
22118469
28004739
20920257
23761445
24845553
+ More
22516182 23537049 28049606 17510324 26369729 28797301 25576852 26830274 24945155 22216098 20966253 18362917 19820115 26108605 12364791 14747013 17210077 20075255 29403074 25244985 20431018 24438588 28648823 21719571 21804562 27762356 22398555 20809919 18464734 10567383 16141072 15489334 21183079 11419946 16365036 28528879 20798317 21282665 21993624
22516182 23537049 28049606 17510324 26369729 28797301 25576852 26830274 24945155 22216098 20966253 18362917 19820115 26108605 12364791 14747013 17210077 20075255 29403074 25244985 20431018 24438588 28648823 21719571 21804562 27762356 22398555 20809919 18464734 10567383 16141072 15489334 21183079 11419946 16365036 28528879 20798317 21282665 21993624
EMBL
KQ459220
KPJ02861.1
ODYU01003368
SOQ42039.1
AGBW02012778
OWR44451.1
+ More
RSAL01000231 RVE43845.1 NWSH01002966 PCG67205.1 KQ459984 KPJ18979.1 GALX01002900 JAB65566.1 AJWK01004132 DS232451 EDS41909.1 GFDL01007208 JAV27837.1 GEZM01101277 JAV52704.1 GGFK01006986 MBW40307.1 ADMH02001019 ETN64391.1 GGFK01006759 MBW40080.1 GGFK01006723 MBW40044.1 GGFK01007069 MBW40390.1 GGFJ01008220 MBW57361.1 GAMD01000937 JAB00654.1 KK852470 KDR23258.1 GGFJ01008002 MBW57143.1 GGFJ01008392 MBW57533.1 CAQQ02110234 CAQQ02110235 GGFJ01008237 MBW57378.1 APGK01017098 BT126918 KB739995 KB630277 KB632201 AEE61880.1 ENN82044.1 ERL83504.1 ERL90053.1 GEFH01002387 JAP66194.1 GGFM01005356 MBW26107.1 BT078578 ACO13002.1 BT121860 ADD38790.1 GBBK01001453 JAC23029.1 GANO01002406 JAB57465.1 CH477396 EAT41838.1 GDAI01002182 JAI15421.1 AXCM01004068 GFPF01012313 MAA23459.1 GACK01008795 JAA56239.1 GBBM01004216 JAC31202.1 GEDV01004077 JAP84480.1 GAPW01003440 JAC10158.1 JO844373 AEO35990.1 GGLE01001587 MBY05713.1 KY031246 ATU82997.1 BT077902 ACO12326.1 GADI01006874 JAA66934.1 APCN01000602 KQ971312 EEZ98443.2 JRES01000091 KNC34204.1 AAAB01008859 EAA07620.4 KYB29167.1 AAZX01007551 PYGN01001697 PSN33329.1 GEFM01003423 JAP72373.1 KQ780345 OAD51950.1 KQ434984 KZC13134.1 CR760766 CAJ83166.1 CCAG010013874 AAMC01097512 AAMC01097513 AAMC01097514 AAMC01097515 AAMC01097516 AAMC01097517 AAMC01097518 AAMC01097519 AAMC01097520 AAMC01097521 ATLV01018673 KE525245 KFB43305.1 NNAY01003086 OXU20005.1 GFWV01020818 MAA45546.1 KQ982649 KYQ53111.1 GL887532 EGI70843.1 GEDC01013845 JAS23453.1 JH000818 EGW12555.1 KQ414710 KOC62986.1 AXCN02001586 CM004476 OCT75986.1 LJIG01001956 KRT84984.1 CABD030055672 CABD030055673 CABD030055674 CABD030055675 CABD030055676 CABD030055677 CABD030055678 CABD030055679 CABD030055680 CABD030055681 AMGL01089547 AMGL01089548 AMGL01089549 AMGL01089550 AMGL01089551 AMGL01089552 AMGL01089553 AMGL01089554 AMGL01089555 AB027568 AK028431 AK035044 AK039131 AK136486 BC015246 BC023354 GFJQ02004274 JAW02696.1 GBYB01012555 GBYB01012558 JAG82322.1 JAG82325.1 GL446946 EFN87047.1 AQIA01048568 AQIA01048569 AQIA01048570 AQIA01048571 AQIA01048572 AQIA01048573 AQIA01048574 GL771642 EFZ09677.1 AAKN02015382 AAKN02015383 AAKN02015384 AAKN02015385 AAKN02015386 AAKN02015387 AGTP01044987 AGTP01044988 AGTP01044989 AGTP01044990 AGTP01044991 AGTP01044992 AGTP01044993 AGTP01044994 AGTP01044995 AGTP01044996 AAPE02029221 AAPE02029222 AAPE02029223 AAPE02029224 AAPE02029225 AAPE02029226
RSAL01000231 RVE43845.1 NWSH01002966 PCG67205.1 KQ459984 KPJ18979.1 GALX01002900 JAB65566.1 AJWK01004132 DS232451 EDS41909.1 GFDL01007208 JAV27837.1 GEZM01101277 JAV52704.1 GGFK01006986 MBW40307.1 ADMH02001019 ETN64391.1 GGFK01006759 MBW40080.1 GGFK01006723 MBW40044.1 GGFK01007069 MBW40390.1 GGFJ01008220 MBW57361.1 GAMD01000937 JAB00654.1 KK852470 KDR23258.1 GGFJ01008002 MBW57143.1 GGFJ01008392 MBW57533.1 CAQQ02110234 CAQQ02110235 GGFJ01008237 MBW57378.1 APGK01017098 BT126918 KB739995 KB630277 KB632201 AEE61880.1 ENN82044.1 ERL83504.1 ERL90053.1 GEFH01002387 JAP66194.1 GGFM01005356 MBW26107.1 BT078578 ACO13002.1 BT121860 ADD38790.1 GBBK01001453 JAC23029.1 GANO01002406 JAB57465.1 CH477396 EAT41838.1 GDAI01002182 JAI15421.1 AXCM01004068 GFPF01012313 MAA23459.1 GACK01008795 JAA56239.1 GBBM01004216 JAC31202.1 GEDV01004077 JAP84480.1 GAPW01003440 JAC10158.1 JO844373 AEO35990.1 GGLE01001587 MBY05713.1 KY031246 ATU82997.1 BT077902 ACO12326.1 GADI01006874 JAA66934.1 APCN01000602 KQ971312 EEZ98443.2 JRES01000091 KNC34204.1 AAAB01008859 EAA07620.4 KYB29167.1 AAZX01007551 PYGN01001697 PSN33329.1 GEFM01003423 JAP72373.1 KQ780345 OAD51950.1 KQ434984 KZC13134.1 CR760766 CAJ83166.1 CCAG010013874 AAMC01097512 AAMC01097513 AAMC01097514 AAMC01097515 AAMC01097516 AAMC01097517 AAMC01097518 AAMC01097519 AAMC01097520 AAMC01097521 ATLV01018673 KE525245 KFB43305.1 NNAY01003086 OXU20005.1 GFWV01020818 MAA45546.1 KQ982649 KYQ53111.1 GL887532 EGI70843.1 GEDC01013845 JAS23453.1 JH000818 EGW12555.1 KQ414710 KOC62986.1 AXCN02001586 CM004476 OCT75986.1 LJIG01001956 KRT84984.1 CABD030055672 CABD030055673 CABD030055674 CABD030055675 CABD030055676 CABD030055677 CABD030055678 CABD030055679 CABD030055680 CABD030055681 AMGL01089547 AMGL01089548 AMGL01089549 AMGL01089550 AMGL01089551 AMGL01089552 AMGL01089553 AMGL01089554 AMGL01089555 AB027568 AK028431 AK035044 AK039131 AK136486 BC015246 BC023354 GFJQ02004274 JAW02696.1 GBYB01012555 GBYB01012558 JAG82322.1 JAG82325.1 GL446946 EFN87047.1 AQIA01048568 AQIA01048569 AQIA01048570 AQIA01048571 AQIA01048572 AQIA01048573 AQIA01048574 GL771642 EFZ09677.1 AAKN02015382 AAKN02015383 AAKN02015384 AAKN02015385 AAKN02015386 AAKN02015387 AGTP01044987 AGTP01044988 AGTP01044989 AGTP01044990 AGTP01044991 AGTP01044992 AGTP01044993 AGTP01044994 AGTP01044995 AGTP01044996 AAPE02029221 AAPE02029222 AAPE02029223 AAPE02029224 AAPE02029225 AAPE02029226
Proteomes
UP000053268
UP000007151
UP000283053
UP000218220
UP000053240
UP000092461
+ More
UP000002320 UP000069272 UP000000673 UP000027135 UP000015102 UP000019118 UP000030742 UP000075900 UP000075881 UP000008820 UP000075883 UP000075903 UP000075920 UP000076407 UP000075902 UP000091820 UP000075840 UP000075882 UP000007266 UP000037069 UP000007062 UP000075885 UP000002358 UP000245037 UP000076502 UP000092445 UP000078200 UP000076408 UP000095300 UP000092444 UP000008143 UP000030765 UP000215335 UP000075880 UP000075901 UP000075809 UP000007755 UP000001075 UP000192223 UP000053825 UP000075886 UP000186698 UP000001519 UP000002356 UP000002279 UP000000589 UP000008237 UP000245340 UP000233100 UP000005447 UP000005215 UP000001074
UP000002320 UP000069272 UP000000673 UP000027135 UP000015102 UP000019118 UP000030742 UP000075900 UP000075881 UP000008820 UP000075883 UP000075903 UP000075920 UP000076407 UP000075902 UP000091820 UP000075840 UP000075882 UP000007266 UP000037069 UP000007062 UP000075885 UP000002358 UP000245037 UP000076502 UP000092445 UP000078200 UP000076408 UP000095300 UP000092444 UP000008143 UP000030765 UP000215335 UP000075880 UP000075901 UP000075809 UP000007755 UP000001075 UP000192223 UP000053825 UP000075886 UP000186698 UP000001519 UP000002356 UP000002279 UP000000589 UP000008237 UP000245340 UP000233100 UP000005447 UP000005215 UP000001074
PRIDE
Pfam
Interpro
IPR006282
Thi_PPkinase
+ More
IPR036759 TPK_catalytic_sf
IPR007373 Thiamin_PyroPKinase_B1-bd
IPR036371 TPK_B1-bd_sf
IPR007371 TPK_catalytic
IPR016966 Thiamin_pyrophosphokinase_euk
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR014748 Enoyl-CoA_hydra_C
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR004364 aa-tRNA-synt_II
IPR004365 NA-bd_OB_tRNA
IPR012340 NA-bd_OB-fold
IPR004522 Asn-tRNA-ligase
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR006195 aa-tRNA-synth_II
IPR036759 TPK_catalytic_sf
IPR007373 Thiamin_PyroPKinase_B1-bd
IPR036371 TPK_B1-bd_sf
IPR007371 TPK_catalytic
IPR016966 Thiamin_pyrophosphokinase_euk
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR014748 Enoyl-CoA_hydra_C
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR004364 aa-tRNA-synt_II
IPR004365 NA-bd_OB_tRNA
IPR012340 NA-bd_OB-fold
IPR004522 Asn-tRNA-ligase
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR006195 aa-tRNA-synth_II
Gene 3D
CDD
ProteinModelPortal
A0A194QHE2
A0A2H1VMH9
A0A212ESN4
A0A3S2LCP0
A0A2A4J547
A0A194RNJ0
+ More
V5I9I0 A0A1B0CAN8 B0X7F9 A0A1Q3FJT3 A0A1Y1JTR1 A0A182FNN9 A0A2M4AHT2 W5JJV2 A0A2M4AH18 A0A2M4AGZ1 A0A2M4AHY5 A0A2M4BWQ1 T1E9W7 A0A067RJS6 A0A2M4BVU1 A0A2M4BWX1 T1GTI5 A0A2M4BXC7 J3JUK3 A0A131XFB5 A0A1Y9HFB6 A0A2M3ZC20 C1BVE9 D3PJQ4 A0A023FP32 A0A182JUP7 U5EJ18 Q175Q4 A0A0K8TMN4 A0A182MFE1 A0A224Z5K2 L7LZG3 A0A023GBB3 A0A131Z148 A0A023ELM5 G3MR72 A0A2R5L890 A0A2K8JX53 C1BTH3 A0A182UUV8 A0A0K8R8Y1 A0A1Y9IVZ1 A0A182WRQ4 A0A182U6J6 A0A1A9WJK7 A0A182HU24 A0A182LG03 D6W952 A0A0L0CS03 Q7QD56 A0A139WMT9 A0A182P6N6 K7J2E2 A0A2P8XMV1 A0A131XYR3 A0A310SEX6 A0A154PNU9 A0A1B0AAB9 A0A1A9UN97 A0A182XWD1 A0A1I8Q7U0 Q28HS1 A0A1B0G6V7 F6TP13 A0A084VZB1 A0A232ENS3 A0A293MYH5 A0A240PNY2 A0A182SMJ9 A0A151WZI5 F4W4E8 A0A1B6DCN3 G3HW81 A0A1W4X2H6 A0A0L7QWN9 A0A182QE81 A0A1L8FWN4 A0A0T6BC94 G3QVB4 W5QAX1 F7EK85 Q9R0M5 A0A1Z5L5K6 A0A0C9RUM1 E2BB74 A0A2U3WL60 A0A2K5VA75 E9JC14 A0A2I2ZCZ0 A0A286X7B8 I3N387 G1P1W6
V5I9I0 A0A1B0CAN8 B0X7F9 A0A1Q3FJT3 A0A1Y1JTR1 A0A182FNN9 A0A2M4AHT2 W5JJV2 A0A2M4AH18 A0A2M4AGZ1 A0A2M4AHY5 A0A2M4BWQ1 T1E9W7 A0A067RJS6 A0A2M4BVU1 A0A2M4BWX1 T1GTI5 A0A2M4BXC7 J3JUK3 A0A131XFB5 A0A1Y9HFB6 A0A2M3ZC20 C1BVE9 D3PJQ4 A0A023FP32 A0A182JUP7 U5EJ18 Q175Q4 A0A0K8TMN4 A0A182MFE1 A0A224Z5K2 L7LZG3 A0A023GBB3 A0A131Z148 A0A023ELM5 G3MR72 A0A2R5L890 A0A2K8JX53 C1BTH3 A0A182UUV8 A0A0K8R8Y1 A0A1Y9IVZ1 A0A182WRQ4 A0A182U6J6 A0A1A9WJK7 A0A182HU24 A0A182LG03 D6W952 A0A0L0CS03 Q7QD56 A0A139WMT9 A0A182P6N6 K7J2E2 A0A2P8XMV1 A0A131XYR3 A0A310SEX6 A0A154PNU9 A0A1B0AAB9 A0A1A9UN97 A0A182XWD1 A0A1I8Q7U0 Q28HS1 A0A1B0G6V7 F6TP13 A0A084VZB1 A0A232ENS3 A0A293MYH5 A0A240PNY2 A0A182SMJ9 A0A151WZI5 F4W4E8 A0A1B6DCN3 G3HW81 A0A1W4X2H6 A0A0L7QWN9 A0A182QE81 A0A1L8FWN4 A0A0T6BC94 G3QVB4 W5QAX1 F7EK85 Q9R0M5 A0A1Z5L5K6 A0A0C9RUM1 E2BB74 A0A2U3WL60 A0A2K5VA75 E9JC14 A0A2I2ZCZ0 A0A286X7B8 I3N387 G1P1W6
PDB
2F17
E-value=6.63104e-48,
Score=479
Ontologies
KEGG
PATHWAY
GO
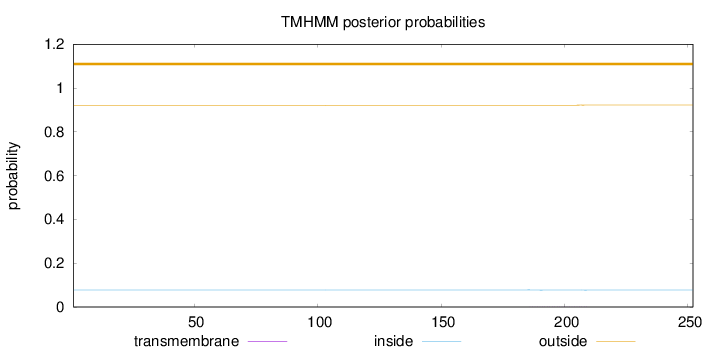
Topology
Length:
252
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01733
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.07807
outside
1 - 252
Population Genetic Test Statistics
Pi
356.496623
Theta
191.806993
Tajima's D
2.082312
CLR
0.129674
CSRT
0.896205189740513
Interpretation
Uncertain
