Gene
KWMTBOMO05210
Pre Gene Modal
BGIBMGA002436
Annotation
PREDICTED:_uncharacterized_protein_LOC106142857_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.388 PlasmaMembrane Reliability : 1.805
Sequence
CDS
ATGTCTAAAGAAATGGCGGCGCCGAAGAGGACTGTAGGCCAGAGAATTGTGGAGGAAATTTACACGTGTTGTGCTTTCTGGATAGCTGCATCCGCTTTACCGGCCTTGGCGGTGTCTTATCTCGTTGTACGTGTGAGCAGACATTTATGGCTTAGACTGCTGTCCAGTAGATATCCAGATTTCGAATTCATCAAAACTGACACAGTCAGAACATTGCTGGACACGCATCGGAATCAGGGGATCATTAACGTACTACTGGCCGTGAAAGGTAATCCGAACGTCGATGAAATCAAAGGAAAACTCATAGAGCACGTTATCGACAAACGAGATAGGAATGGACAGTTCATGTTTCCAAGGTTACGACATTTACTCGTCTCATGCTGGGGCAACTACGCTTGGGATGTCAACGTGCGTTTTCGGTTTGAAAACCACTTCATCGTAGCTAATGCTGTGTACAGAGGCCGGCCGGTTACAGAGAGTAATATACAGGAATACATCAGTGACATCGTATCAAAATATTTCTCTCCAGACCAACCGCCCTGGCAATACATAATAATACCGTGTGCAGCTACAGAACCAAAATATTATATTTTAGTTCGCGTGCACCATCTGCTACTATCCGGCGCGAAGTCTATTAACATCGGAGATTTATTACTGGTCGAGCAATTGAAACAAACAGATAGAATGGCTCAAGAATATACACAACAAAGCCCTTTAACGAAATTATTTCCAACGCCTTCTGCCATACCTCAATTGTGGGAAAAGCTTCATGAAAATATGTCGAATACATGGAACGAATTCGTTTCTGAATACGACCCCGTCGAAAGCCCTCGTGCTTTGAAAAGCATGCCGGGTGCGTTCCACGTCGCCGGTCTGTTATTGATATCCAGCGTGAGCGCTTTGAGAGAACTCAACAAGAAATCGAATGGCAGAAACAGAGCCACTGATGGTCCGCTCACAGCAACTACTCTACTTGCAGCAGTGAGAAGAGAATGTAAAAGGAGAAGCTTAACTATAATTAAAGTTTTCTTGTCACCGCTAGTAACAGCAGACCCGAGAAAATGGCCTAAAAGAACATTTAAGACTATCACGACATCGACTGTTTTAATGTTGAAATTACCGTTTCGCGTTCGCGATGAAATTTTAGCTTTAAACGAACTAAAGAATACTGGTCACGTAAAACAGCCTAACACTTTAACGTGGAGGTACGGGGAATTAGCGCAGCTTTGCGTGAGAGCTACTAATGAAGCGTGGAATGGCTTACTAGAAATTTATAGGGCTCCAGCAAAATTGTGGACGGACACCATTAGAACGGACGATGGGAAGCGACACTTGTTACAAGCCGTTTCTCTGTGCGGACGGAAGGTTATATCATGGTCTCGTCCCGTATCCCGAACCGCAATCGAACGCGTGGCTCGGGCGTTGGGCGTATCATCGACAGACGTCGTTTTGTTCGCCGCAACGGATGCCTTACGAGCATTCTTTGAACAAGCACACGGGGACCCGCCTGACATAGTACTCACAACAGCTAGGGCCGCTTCAGAAGATTTCCTATTCACTTTCGCGGAGGGCCAGGGGAAGATGTATAAAAAATCACAAACTGGTGGTATGGTCTGTCTGACCCTGCCTATAGGCGCGAGTCCACGTCGTATCGCCGCCGTAGTGGAACAAGCGTGCAATAGACAGCAGGCATTGGCCGGGGCCTGGGCGGCCCAGGCTCGATGCGGTGCATTAACTAGAGCTGTTCCTTCTCCTCTCGCACGATTAGCATTGAATATACTGTCAAGAAGATACGCAGTGTCTTATGCGGAAATCAACGCACCTCTCAATGCCAAACAACGGGCCACACTGTGGGGTCAGACTGTCGATCACGTTATCTATTGGCGCCCGCCTCAGGCCAATATCAGTATGTCACTAACAGTGATACAATACGGGGAGACGGTGAGGCTGGCCGTGATGGCGGACGCGAGGCTGTCTCCGTCACATTCCGCCCCCACCACGCGATGGCCCGTCGCAGTCGAGCAACTAATCAATAAAGTCGATACGGAAATATCTCGCATAGCCACAACACTACAGCCCGACATCCCTACTGCGGTCGAAACTCGAGAAGCTGACGACACTTACGAGGCAGAAACGTCGCGTGACGTCACCAGCTCGAGCGGACTGAGGCCGCCGGTTTTGACAGCTGTCAGTCCGCCGCCATTGCGACGCAGAATGACGCACCACTGA
Protein
MSKEMAAPKRTVGQRIVEEIYTCCAFWIAASALPALAVSYLVVRVSRHLWLRLLSSRYPDFEFIKTDTVRTLLDTHRNQGIINVLLAVKGNPNVDEIKGKLIEHVIDKRDRNGQFMFPRLRHLLVSCWGNYAWDVNVRFRFENHFIVANAVYRGRPVTESNIQEYISDIVSKYFSPDQPPWQYIIIPCAATEPKYYILVRVHHLLLSGAKSINIGDLLLVEQLKQTDRMAQEYTQQSPLTKLFPTPSAIPQLWEKLHENMSNTWNEFVSEYDPVESPRALKSMPGAFHVAGLLLISSVSALRELNKKSNGRNRATDGPLTATTLLAAVRRECKRRSLTIIKVFLSPLVTADPRKWPKRTFKTITTSTVLMLKLPFRVRDEILALNELKNTGHVKQPNTLTWRYGELAQLCVRATNEAWNGLLEIYRAPAKLWTDTIRTDDGKRHLLQAVSLCGRKVISWSRPVSRTAIERVARALGVSSTDVVLFAATDALRAFFEQAHGDPPDIVLTTARAASEDFLFTFAEGQGKMYKKSQTGGMVCLTLPIGASPRRIAAVVEQACNRQQALAGAWAAQARCGALTRAVPSPLARLALNILSRRYAVSYAEINAPLNAKQRATLWGQTVDHVIYWRPPQANISMSLTVIQYGETVRLAVMADARLSPSHSAPTTRWPVAVEQLINKVDTEISRIATTLQPDIPTAVETREADDTYEAETSRDVTSSSGLRPPVLTAVSPPPLRRRMTHH
Summary
Uniprot
A0A194QBD5
A0A2H1VR50
A0A212ESI7
A0A0L7L268
A0A194RN82
D6WW58
+ More
A0A067QWP0 A0A1B6MLY5 U5ES27 A0A1Q3FMK7 B0W731 E0VFL2 A0A1Q3FLJ4 J3JU66 U4U357 N6T9H5 A0A2J7QWR0 A0A0A9W240 A0A1Y1ML89 A0A2P8Y963 A0A1W4WSB7 R4FLA5 A0A0T6BE80 A0A1W4WR06 A0A1L8DA55 A0A2J7QWQ7 A0A182GS77 A0A087ZZI5 A0A0K8V616 A0A0A1WI32 A0A1B0EUF8 W8BVW9 A0A1L8D9H6 A0A034VET3 A0A154PJE2 A0A0L7QNC1 A0A310SEE8 A0A1W4V9G7 A0A0L0BZT3 B3MUE8 A0A026WKI8 A0A1I8NB57 A0A3Q0IQM9 Q9VJ41 B4JBK4 A0A1B6FGI0 D3TNF3 A0A0M4ERB9 E2BIZ6 B4P9Z2 A0A1A9WXV5 A0A2A3EM21 B4LUH3 Q29MY8 B4G8I7 E9JCL5 A0A1B0AV44 A0A1A9Y5M8 A0A1I8PJS1 A0A3B0JU31 A0A0J9R590 B3NL87 B4KHY8 A0A0M9A6A1 K7ISI0 A0A158NQA5 A0A1B6C8K1 A0A151JNY5 B4MTS7 A0A336LT33 A0A232EYN5 A0A1B0G0X5 E2B034 A0A1A9ZPS6 A0A1A9VJC5 T1PC59 A0A195B5S7 A0A2S2P3W4 F4WBF4 X1XR31
A0A067QWP0 A0A1B6MLY5 U5ES27 A0A1Q3FMK7 B0W731 E0VFL2 A0A1Q3FLJ4 J3JU66 U4U357 N6T9H5 A0A2J7QWR0 A0A0A9W240 A0A1Y1ML89 A0A2P8Y963 A0A1W4WSB7 R4FLA5 A0A0T6BE80 A0A1W4WR06 A0A1L8DA55 A0A2J7QWQ7 A0A182GS77 A0A087ZZI5 A0A0K8V616 A0A0A1WI32 A0A1B0EUF8 W8BVW9 A0A1L8D9H6 A0A034VET3 A0A154PJE2 A0A0L7QNC1 A0A310SEE8 A0A1W4V9G7 A0A0L0BZT3 B3MUE8 A0A026WKI8 A0A1I8NB57 A0A3Q0IQM9 Q9VJ41 B4JBK4 A0A1B6FGI0 D3TNF3 A0A0M4ERB9 E2BIZ6 B4P9Z2 A0A1A9WXV5 A0A2A3EM21 B4LUH3 Q29MY8 B4G8I7 E9JCL5 A0A1B0AV44 A0A1A9Y5M8 A0A1I8PJS1 A0A3B0JU31 A0A0J9R590 B3NL87 B4KHY8 A0A0M9A6A1 K7ISI0 A0A158NQA5 A0A1B6C8K1 A0A151JNY5 B4MTS7 A0A336LT33 A0A232EYN5 A0A1B0G0X5 E2B034 A0A1A9ZPS6 A0A1A9VJC5 T1PC59 A0A195B5S7 A0A2S2P3W4 F4WBF4 X1XR31
Pubmed
26354079
22118469
26227816
18362917
19820115
24845553
+ More
20566863 22516182 23537049 25401762 26823975 28004739 29403074 26483478 25830018 24495485 25348373 26108605 17994087 24508170 30249741 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20798317 17550304 15632085 21282665 22936249 20075255 21347285 28648823 21719571
20566863 22516182 23537049 25401762 26823975 28004739 29403074 26483478 25830018 24495485 25348373 26108605 17994087 24508170 30249741 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20798317 17550304 15632085 21282665 22936249 20075255 21347285 28648823 21719571
EMBL
KQ459220
KPJ02853.1
ODYU01003946
SOQ43321.1
AGBW02012778
OWR44458.1
+ More
JTDY01003519 KOB69421.1 KQ459984 KPJ18972.1 KQ971361 EFA08670.2 KK853278 KDR09061.1 GEBQ01003019 JAT36958.1 GANO01005001 JAB54870.1 GFDL01006329 JAV28716.1 DS231851 EDS37305.1 DS235115 EEB12168.1 GFDL01006657 JAV28388.1 BT126779 AEE61741.1 KB631578 ERL84405.1 APGK01038972 APGK01038973 KB740966 ENN76909.1 NEVH01009422 PNF33022.1 GBHO01043091 GBHO01043089 GBRD01013308 GDHC01009139 JAG00513.1 JAG00515.1 JAG52518.1 JAQ09490.1 GEZM01028107 JAV86461.1 PYGN01000790 PSN40792.1 ACPB03000196 GAHY01002174 JAA75336.1 LJIG01001289 KRT85651.1 GFDF01010864 JAV03220.1 PNF33020.1 JXUM01016949 KQ560470 KXJ82251.1 GDHF01018013 JAI34301.1 GBXI01015593 JAC98698.1 AJWK01015077 AJWK01015078 AJWK01015079 GAMC01005487 JAC01069.1 GFDF01010955 JAV03129.1 GAKP01017146 JAC41806.1 KQ434924 KZC11584.1 KQ414855 KOC60125.1 KQ779356 OAD51998.1 JRES01001109 KNC25496.1 CH902624 EDV33477.1 KK107167 QOIP01000010 EZA56498.1 RLU17783.1 AE014134 AY058338 AAF53716.2 AAL13567.1 CH916368 EDW04027.1 GECZ01020458 JAS49311.1 EZ422955 ADD19231.1 CP012523 ALC39407.1 GL448543 EFN84341.1 CM000158 EDW90333.2 KZ288212 PBC32805.1 CH940649 EDW64159.1 CH379060 EAL33555.2 CH479180 EDW28667.1 GL771849 EFZ09428.1 JXJN01003956 OUUW01000004 SPP78980.1 CM002910 KMY90889.1 CH954179 EDV54737.2 CH933807 EDW11270.1 KQ435732 KOX77336.1 ADTU01023000 ADTU01023001 GEDC01027723 JAS09575.1 KQ978820 KYN28132.1 CH963852 EDW75516.1 UFQT01000177 SSX21186.1 NNAY01001665 OXU23278.1 CCAG010001389 GL444372 EFN60960.1 KA646274 AFP60903.1 KQ976595 KYM79625.1 GGMR01011491 MBY24110.1 GL888062 EGI68510.1 ABLF02031390 ABLF02037396
JTDY01003519 KOB69421.1 KQ459984 KPJ18972.1 KQ971361 EFA08670.2 KK853278 KDR09061.1 GEBQ01003019 JAT36958.1 GANO01005001 JAB54870.1 GFDL01006329 JAV28716.1 DS231851 EDS37305.1 DS235115 EEB12168.1 GFDL01006657 JAV28388.1 BT126779 AEE61741.1 KB631578 ERL84405.1 APGK01038972 APGK01038973 KB740966 ENN76909.1 NEVH01009422 PNF33022.1 GBHO01043091 GBHO01043089 GBRD01013308 GDHC01009139 JAG00513.1 JAG00515.1 JAG52518.1 JAQ09490.1 GEZM01028107 JAV86461.1 PYGN01000790 PSN40792.1 ACPB03000196 GAHY01002174 JAA75336.1 LJIG01001289 KRT85651.1 GFDF01010864 JAV03220.1 PNF33020.1 JXUM01016949 KQ560470 KXJ82251.1 GDHF01018013 JAI34301.1 GBXI01015593 JAC98698.1 AJWK01015077 AJWK01015078 AJWK01015079 GAMC01005487 JAC01069.1 GFDF01010955 JAV03129.1 GAKP01017146 JAC41806.1 KQ434924 KZC11584.1 KQ414855 KOC60125.1 KQ779356 OAD51998.1 JRES01001109 KNC25496.1 CH902624 EDV33477.1 KK107167 QOIP01000010 EZA56498.1 RLU17783.1 AE014134 AY058338 AAF53716.2 AAL13567.1 CH916368 EDW04027.1 GECZ01020458 JAS49311.1 EZ422955 ADD19231.1 CP012523 ALC39407.1 GL448543 EFN84341.1 CM000158 EDW90333.2 KZ288212 PBC32805.1 CH940649 EDW64159.1 CH379060 EAL33555.2 CH479180 EDW28667.1 GL771849 EFZ09428.1 JXJN01003956 OUUW01000004 SPP78980.1 CM002910 KMY90889.1 CH954179 EDV54737.2 CH933807 EDW11270.1 KQ435732 KOX77336.1 ADTU01023000 ADTU01023001 GEDC01027723 JAS09575.1 KQ978820 KYN28132.1 CH963852 EDW75516.1 UFQT01000177 SSX21186.1 NNAY01001665 OXU23278.1 CCAG010001389 GL444372 EFN60960.1 KA646274 AFP60903.1 KQ976595 KYM79625.1 GGMR01011491 MBY24110.1 GL888062 EGI68510.1 ABLF02031390 ABLF02037396
Proteomes
UP000053268
UP000007151
UP000037510
UP000053240
UP000007266
UP000027135
+ More
UP000002320 UP000009046 UP000030742 UP000019118 UP000235965 UP000245037 UP000192223 UP000015103 UP000069940 UP000249989 UP000005203 UP000092461 UP000076502 UP000053825 UP000192221 UP000037069 UP000007801 UP000053097 UP000279307 UP000095301 UP000079169 UP000000803 UP000001070 UP000092553 UP000008237 UP000002282 UP000091820 UP000242457 UP000008792 UP000001819 UP000008744 UP000092460 UP000092443 UP000095300 UP000268350 UP000008711 UP000009192 UP000053105 UP000002358 UP000005205 UP000078492 UP000007798 UP000215335 UP000092444 UP000000311 UP000092445 UP000078200 UP000078540 UP000007755 UP000007819
UP000002320 UP000009046 UP000030742 UP000019118 UP000235965 UP000245037 UP000192223 UP000015103 UP000069940 UP000249989 UP000005203 UP000092461 UP000076502 UP000053825 UP000192221 UP000037069 UP000007801 UP000053097 UP000279307 UP000095301 UP000079169 UP000000803 UP000001070 UP000092553 UP000008237 UP000002282 UP000091820 UP000242457 UP000008792 UP000001819 UP000008744 UP000092460 UP000092443 UP000095300 UP000268350 UP000008711 UP000009192 UP000053105 UP000002358 UP000005205 UP000078492 UP000007798 UP000215335 UP000092444 UP000000311 UP000092445 UP000078200 UP000078540 UP000007755 UP000007819
PRIDE
Pfam
Interpro
IPR009721
O-acyltransferase_WSD1_C
+ More
IPR019354 Smg8/Smg9
IPR028802 SMG8
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR040240 TAF1
IPR036427 Bromodomain-like_sf
IPR036741 TAFII-230_TBP-bd_sf
IPR041670 Znf-CCHC_6
IPR009067 TAF_II_230-bd
IPR022591 TFIID_sub1_DUF3591
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
IPR019354 Smg8/Smg9
IPR028802 SMG8
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR040240 TAF1
IPR036427 Bromodomain-like_sf
IPR036741 TAFII-230_TBP-bd_sf
IPR041670 Znf-CCHC_6
IPR009067 TAF_II_230-bd
IPR022591 TFIID_sub1_DUF3591
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
Gene 3D
ProteinModelPortal
A0A194QBD5
A0A2H1VR50
A0A212ESI7
A0A0L7L268
A0A194RN82
D6WW58
+ More
A0A067QWP0 A0A1B6MLY5 U5ES27 A0A1Q3FMK7 B0W731 E0VFL2 A0A1Q3FLJ4 J3JU66 U4U357 N6T9H5 A0A2J7QWR0 A0A0A9W240 A0A1Y1ML89 A0A2P8Y963 A0A1W4WSB7 R4FLA5 A0A0T6BE80 A0A1W4WR06 A0A1L8DA55 A0A2J7QWQ7 A0A182GS77 A0A087ZZI5 A0A0K8V616 A0A0A1WI32 A0A1B0EUF8 W8BVW9 A0A1L8D9H6 A0A034VET3 A0A154PJE2 A0A0L7QNC1 A0A310SEE8 A0A1W4V9G7 A0A0L0BZT3 B3MUE8 A0A026WKI8 A0A1I8NB57 A0A3Q0IQM9 Q9VJ41 B4JBK4 A0A1B6FGI0 D3TNF3 A0A0M4ERB9 E2BIZ6 B4P9Z2 A0A1A9WXV5 A0A2A3EM21 B4LUH3 Q29MY8 B4G8I7 E9JCL5 A0A1B0AV44 A0A1A9Y5M8 A0A1I8PJS1 A0A3B0JU31 A0A0J9R590 B3NL87 B4KHY8 A0A0M9A6A1 K7ISI0 A0A158NQA5 A0A1B6C8K1 A0A151JNY5 B4MTS7 A0A336LT33 A0A232EYN5 A0A1B0G0X5 E2B034 A0A1A9ZPS6 A0A1A9VJC5 T1PC59 A0A195B5S7 A0A2S2P3W4 F4WBF4 X1XR31
A0A067QWP0 A0A1B6MLY5 U5ES27 A0A1Q3FMK7 B0W731 E0VFL2 A0A1Q3FLJ4 J3JU66 U4U357 N6T9H5 A0A2J7QWR0 A0A0A9W240 A0A1Y1ML89 A0A2P8Y963 A0A1W4WSB7 R4FLA5 A0A0T6BE80 A0A1W4WR06 A0A1L8DA55 A0A2J7QWQ7 A0A182GS77 A0A087ZZI5 A0A0K8V616 A0A0A1WI32 A0A1B0EUF8 W8BVW9 A0A1L8D9H6 A0A034VET3 A0A154PJE2 A0A0L7QNC1 A0A310SEE8 A0A1W4V9G7 A0A0L0BZT3 B3MUE8 A0A026WKI8 A0A1I8NB57 A0A3Q0IQM9 Q9VJ41 B4JBK4 A0A1B6FGI0 D3TNF3 A0A0M4ERB9 E2BIZ6 B4P9Z2 A0A1A9WXV5 A0A2A3EM21 B4LUH3 Q29MY8 B4G8I7 E9JCL5 A0A1B0AV44 A0A1A9Y5M8 A0A1I8PJS1 A0A3B0JU31 A0A0J9R590 B3NL87 B4KHY8 A0A0M9A6A1 K7ISI0 A0A158NQA5 A0A1B6C8K1 A0A151JNY5 B4MTS7 A0A336LT33 A0A232EYN5 A0A1B0G0X5 E2B034 A0A1A9ZPS6 A0A1A9VJC5 T1PC59 A0A195B5S7 A0A2S2P3W4 F4WBF4 X1XR31
Ontologies
GO
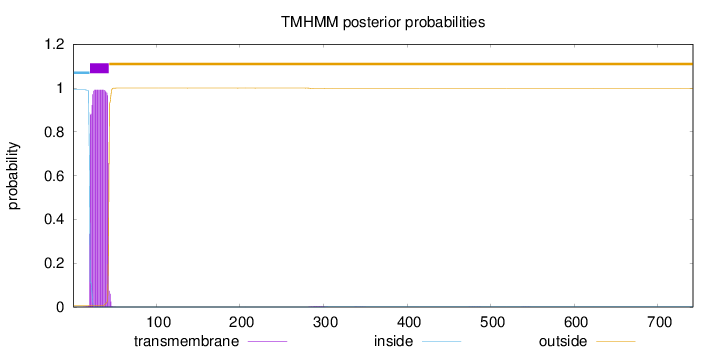
Topology
Length:
742
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.6785899999999
Exp number, first 60 AAs:
22.62068
Total prob of N-in:
0.99213
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 742
Population Genetic Test Statistics
Pi
201.556199
Theta
172.627492
Tajima's D
0.587426
CLR
0.16377
CSRT
0.547322633868307
Interpretation
Possibly Positive selection
