Gene
KWMTBOMO05205 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002462
Annotation
PREDICTED:_bifunctional_purine_biosynthesis_protein_PURH_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.763
Sequence
CDS
ATGGCGTCAAATGGAAAACTAGCTCTTCTCAGCGTTTCAGACAAGACGGGTCTACTCTCGTTAGCAAAGAGCCTGTCGGAATGTGGCCTGCAGTTGATTGCCAGTGGCGGTACCGCCACGGCGCTTCGGAACGCCGGCCTCACAGTTCAAGATGTGTCGGACATCACGAGAGCACCGGAGATGCTCGGAGGTCGGGTGAAAACTTTACATCCAGCGGTACATGCTGGGATCTTAGCTCGATTATCCGACTCTGACCAGGAAGACATGAAACGTCAGAAGTACGAGATGATAAGCGTGGTGGTCTGTAACCTGTACCCGTTCGTCCAGACGGTATCTAAGCCGGACGTGACCGTGGCGGACGCGGTCGAGAACATCGACATCGGCGGCGTGACCCTCCTGCGCGCAGCAGCCAAGAACCACGACCGGGTCACCGTCGTCTGTGACCCGGCCGACTACGATGCTGTAGTCAAAGAAATCAAAGAGAACAAACATCATCAGACGACTTTGGGCACAAGGCAGAGATTAGCCCTGAAGGCGTTCACTCATACTTCGGACTATGACCTCGCCATATCGGACTACTTCCGCAAGCAATACTCGCCCGGGCAAGCCCAACTGACCTTAAGATACGGTATGAACCCACATCAGAAGCCGGCCCAGGTATTCACGACCAGAGACAGCTTGCCGATCACGACACTGAACGGTGCGCCTGGATTTATCAACCTGTGCGACGCGCTGAACGCCTGGCAGCTCGTCAAGGAGCTGAAGGAAGCGCTGAGCCTCCCTGCTGCGGCTAGCTTCAAGCACGTGTCCCCTGCCGGAGCTGCCGTGGGCTTGCCACTCACGGATGAAGAGGCGGCGGTGTGCATGGTGGCGGGCGAACTAAGCGCGCTGGCGTGCGCGTACGCGCGGGCTCGCGGCGCCGACCGCATGAGCTCCTTCGGGGACTTCGTGGCGCTCTCCGACCCCTGTGACGTCAGCACCGCCACCATCATCTCCAGGGAGGTGTCCGACGGCGTCATCGCGCCGGGATACTCGCCCGAAGCACTCAAACTACTCAGCAAGAAAAAGGGCGGGAACTACTGTGTTTTAAAGATCGATCCCACTTACGAACCGAGTCTCATGGAGCAAAAGACCATCTTCGGTTTAACATTGGAGCAAAAACGTAACGACGCGAAGATCACCGCTGAACTTTTCAAGAACGTCGTGACCACCAAGAAGGATTTGCCATCGAACGCCGTGAGGGACCTCATCGTGGCGACCATTGCTCTAAAATACACTCAGAGTAACTCGGTCTGCTTCGCCCGTGATGGACAGGTGATCGGCATCGGTGCGGGGCAGCAGTCGCGCATCCACTGCACGCGGCTGGCGGGGGGCAAGGCGGCGCTGTGGTGGCTGCGCCGACACCCCAGCGTGCTCGCCATGAGGTTCAGGCAGGGCGTCACGCGCGCGGTGCAGGCCAACGCCATCGACAACTACGTCAACGGAACTGTCGGATCTGATTTGCCATTAGAACAATGGGACACGTTGTTCGAAGGCAAACCACCAGCCCTATTCACGGACTCCCAACGCGAGGAATGGATCAAGAAGATGGATAAAGTGGCCCTCGCTTCCGACGCTTTCTTCCCGTTCAGAGACAACATCGACCGAGCTGTGCAGTGCGGCGTGGAATACATCGGCAGTCCGTCTGGTTCGAACAACGATCAAGAAGTCATCGAAGCGTGCAACGAACACAAAATAGCGCTCGCGCATACTAACTTGCGTCTTTTCCACCATTGA
Protein
MASNGKLALLSVSDKTGLLSLAKSLSECGLQLIASGGTATALRNAGLTVQDVSDITRAPEMLGGRVKTLHPAVHAGILARLSDSDQEDMKRQKYEMISVVVCNLYPFVQTVSKPDVTVADAVENIDIGGVTLLRAAAKNHDRVTVVCDPADYDAVVKEIKENKHHQTTLGTRQRLALKAFTHTSDYDLAISDYFRKQYSPGQAQLTLRYGMNPHQKPAQVFTTRDSLPITTLNGAPGFINLCDALNAWQLVKELKEALSLPAAASFKHVSPAGAAVGLPLTDEEAAVCMVAGELSALACAYARARGADRMSSFGDFVALSDPCDVSTATIISREVSDGVIAPGYSPEALKLLSKKKGGNYCVLKIDPTYEPSLMEQKTIFGLTLEQKRNDAKITAELFKNVVTTKKDLPSNAVRDLIVATIALKYTQSNSVCFARDGQVIGIGAGQQSRIHCTRLAGGKAALWWLRRHPSVLAMRFRQGVTRAVQANAIDNYVNGTVGSDLPLEQWDTLFEGKPPALFTDSQREEWIKKMDKVALASDAFFPFRDNIDRAVQCGVEYIGSPSGSNNDQEVIEACNEHKIALAHTNLRLFHH
Summary
Uniprot
H9IYX7
A0A2H1W273
A0A212ESJ3
A0A3S2TDI0
A0A194QCY9
A0A1B6DMZ0
+ More
D6WCW8 A0A1I8PIY0 A0A1D2NDW5 A0A067R8I0 A0A1B0C4G9 A0A1A9ZGE8 A0A1A9XDJ0 A0A1A9WSH0 A0A1A9VJE5 A0A1I8M3Z4 A0A2J7QZU7 D3TNS1 T1PF22 A0A0L0CQM5 A0A034VMJ3 A0A2A4J3H0 B4KBR8 A0A0K8SEJ0 A0A026VYF2 A0A226DFH0 A0A336M7L1 A0A336M7E7 A0A161M7T9 W8ARC0 A0A0P4VXM9 R4G4X2 Q16KZ1 A0A3L8D3G3 A0A1Y1M8J1 A0A1S4FX57 A0A3L8D4H0 A0A2T7PRQ9 A0A182T9B2 A0A023F8J9 K7IY66 A0A182LWN9 A0A182VYT2 A0A1W4W2D2 B4PV20 A0A1B0DCX6 A0A182Q0F1 A0A1Q3F4L1 Q9VC18 B4M648 E9ILL7 B4NI49 B0W5Z3 B3P6W1 A0A182RQ07 B4QTV0 A0A2C9GQJ3 A0A023EV17 A0A182VMZ0 A0A195CI10 B3LX08 A0A182UA66 A0A069DVW4 A0A232FCX1 A0A0V0G555 A0A0P9A1E3 Q7PXI8 B4JYA9 B4HHN0 A0A182N5G2 A0A0M4EQV3 Q6UZ77 A0A182IMK8 A0A195EMX5 A0A3B0K9U7 Q29A03 N6UNV6 B4G3K2 A0A0J7KUY0 A0A131XHL2 E2BQ40 A0A151XEX3 A0A154PBZ5 F4WFC1 A0A2M4BGS7 A0A182X9W5 A0A224Z659 A0A195F9N9 A0A2M4BGG0 E2A229 A0A2M3Z2V6 A0A2M4BHE0 V5IJL0 A0A131YVR5 A0A2M4BGK0 E0V9D9 L7MGB9 U4U9G6 A0A158NCN7 A0A2M4AFR0
D6WCW8 A0A1I8PIY0 A0A1D2NDW5 A0A067R8I0 A0A1B0C4G9 A0A1A9ZGE8 A0A1A9XDJ0 A0A1A9WSH0 A0A1A9VJE5 A0A1I8M3Z4 A0A2J7QZU7 D3TNS1 T1PF22 A0A0L0CQM5 A0A034VMJ3 A0A2A4J3H0 B4KBR8 A0A0K8SEJ0 A0A026VYF2 A0A226DFH0 A0A336M7L1 A0A336M7E7 A0A161M7T9 W8ARC0 A0A0P4VXM9 R4G4X2 Q16KZ1 A0A3L8D3G3 A0A1Y1M8J1 A0A1S4FX57 A0A3L8D4H0 A0A2T7PRQ9 A0A182T9B2 A0A023F8J9 K7IY66 A0A182LWN9 A0A182VYT2 A0A1W4W2D2 B4PV20 A0A1B0DCX6 A0A182Q0F1 A0A1Q3F4L1 Q9VC18 B4M648 E9ILL7 B4NI49 B0W5Z3 B3P6W1 A0A182RQ07 B4QTV0 A0A2C9GQJ3 A0A023EV17 A0A182VMZ0 A0A195CI10 B3LX08 A0A182UA66 A0A069DVW4 A0A232FCX1 A0A0V0G555 A0A0P9A1E3 Q7PXI8 B4JYA9 B4HHN0 A0A182N5G2 A0A0M4EQV3 Q6UZ77 A0A182IMK8 A0A195EMX5 A0A3B0K9U7 Q29A03 N6UNV6 B4G3K2 A0A0J7KUY0 A0A131XHL2 E2BQ40 A0A151XEX3 A0A154PBZ5 F4WFC1 A0A2M4BGS7 A0A182X9W5 A0A224Z659 A0A195F9N9 A0A2M4BGG0 E2A229 A0A2M3Z2V6 A0A2M4BHE0 V5IJL0 A0A131YVR5 A0A2M4BGK0 E0V9D9 L7MGB9 U4U9G6 A0A158NCN7 A0A2M4AFR0
Pubmed
19121390
22118469
26354079
18362917
19820115
27289101
+ More
24845553 25315136 20353571 26108605 25348373 17994087 24508170 24495485 27129103 17510324 30249741 28004739 25474469 20075255 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24945155 26334808 28648823 12364791 14747013 17210077 15632085 23537049 28049606 20798317 21719571 28797301 25765539 26830274 20566863 25576852 21347285
24845553 25315136 20353571 26108605 25348373 17994087 24508170 24495485 27129103 17510324 30249741 28004739 25474469 20075255 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24945155 26334808 28648823 12364791 14747013 17210077 15632085 23537049 28049606 20798317 21719571 28797301 25765539 26830274 20566863 25576852 21347285
EMBL
BABH01010717
ODYU01005802
SOQ47056.1
AGBW02012778
OWR44465.1
RSAL01000293
+ More
RVE42873.1 KQ459220 KPJ02845.1 GEDC01010269 JAS27029.1 KQ971311 EEZ98830.1 LJIJ01000073 ODN03454.1 KK852634 KDR19778.1 JXJN01025444 NEVH01009067 PNF34103.1 EZ423073 ADD19349.1 KA647397 AFP62026.1 JRES01000062 KNC34482.1 GAKP01014396 JAC44556.1 NWSH01003667 PCG65952.1 CH933806 EDW14745.1 GBRD01014113 JAG51713.1 KK107609 EZA48491.1 LNIX01000021 OXA43728.1 UFQT01000655 SSX26195.1 SSX26196.1 GEMB01000566 JAS02564.1 GAMC01019437 JAB87118.1 GDKW01001175 JAI55420.1 ACPB03008077 GAHY01001000 JAA76510.1 CH477930 EAT34980.1 QOIP01000014 RLU14930.1 GEZM01041009 JAV80655.1 RLU15031.1 PZQS01000002 PVD36112.1 GBBI01001037 JAC17675.1 AXCM01003232 CM000160 EDW98869.1 AJVK01031469 AJVK01031470 AXCN02001172 GFDL01012586 JAV22459.1 AE014297 BT011462 AAF56358.1 AAR99120.1 CH940652 EDW59124.2 GL764129 EFZ18470.1 CH964272 EDW84741.1 DS231845 EDS35980.1 CH954182 EDV53781.1 CM000364 EDX14304.1 APCN01000869 GAPW01000650 JAC12948.1 KQ977791 KYM99713.1 CH902617 EDV43847.1 GBGD01000923 JAC87966.1 NNAY01000399 OXU28656.1 GECL01002922 JAP03202.1 KPU80502.1 AAAB01008987 EAA01766.4 CH916377 EDV90671.1 CH480815 EDW43572.1 CP012526 ALC46785.1 AY357301 AAQ63487.1 KQ978691 KYN29252.1 OUUW01000005 SPP80328.1 CM000070 EAL27548.2 APGK01024356 APGK01024357 KB740544 ENN80417.1 CH479179 EDW25010.1 LBMM01002849 KMQ94287.1 GEFH01001978 JAP66603.1 GL449694 EFN82228.1 KQ982215 KYQ58944.1 KQ434869 KZC09416.1 GL888115 EGI67172.1 GGFJ01003115 MBW52256.1 GFPF01010614 MAA21760.1 KQ981727 KYN36927.1 GGFJ01003005 MBW52146.1 GL435917 EFN72513.1 GGFM01002103 MBW22854.1 GGFJ01003077 MBW52218.1 GANP01001252 JAB83216.1 GEDV01005839 JAP82718.1 GGFJ01003026 MBW52167.1 DS234992 EEB09995.1 GACK01002810 JAA62224.1 KB632130 ERL88968.1 ADTU01012001 ADTU01012002 ADTU01012003 GGFK01006292 MBW39613.1
RVE42873.1 KQ459220 KPJ02845.1 GEDC01010269 JAS27029.1 KQ971311 EEZ98830.1 LJIJ01000073 ODN03454.1 KK852634 KDR19778.1 JXJN01025444 NEVH01009067 PNF34103.1 EZ423073 ADD19349.1 KA647397 AFP62026.1 JRES01000062 KNC34482.1 GAKP01014396 JAC44556.1 NWSH01003667 PCG65952.1 CH933806 EDW14745.1 GBRD01014113 JAG51713.1 KK107609 EZA48491.1 LNIX01000021 OXA43728.1 UFQT01000655 SSX26195.1 SSX26196.1 GEMB01000566 JAS02564.1 GAMC01019437 JAB87118.1 GDKW01001175 JAI55420.1 ACPB03008077 GAHY01001000 JAA76510.1 CH477930 EAT34980.1 QOIP01000014 RLU14930.1 GEZM01041009 JAV80655.1 RLU15031.1 PZQS01000002 PVD36112.1 GBBI01001037 JAC17675.1 AXCM01003232 CM000160 EDW98869.1 AJVK01031469 AJVK01031470 AXCN02001172 GFDL01012586 JAV22459.1 AE014297 BT011462 AAF56358.1 AAR99120.1 CH940652 EDW59124.2 GL764129 EFZ18470.1 CH964272 EDW84741.1 DS231845 EDS35980.1 CH954182 EDV53781.1 CM000364 EDX14304.1 APCN01000869 GAPW01000650 JAC12948.1 KQ977791 KYM99713.1 CH902617 EDV43847.1 GBGD01000923 JAC87966.1 NNAY01000399 OXU28656.1 GECL01002922 JAP03202.1 KPU80502.1 AAAB01008987 EAA01766.4 CH916377 EDV90671.1 CH480815 EDW43572.1 CP012526 ALC46785.1 AY357301 AAQ63487.1 KQ978691 KYN29252.1 OUUW01000005 SPP80328.1 CM000070 EAL27548.2 APGK01024356 APGK01024357 KB740544 ENN80417.1 CH479179 EDW25010.1 LBMM01002849 KMQ94287.1 GEFH01001978 JAP66603.1 GL449694 EFN82228.1 KQ982215 KYQ58944.1 KQ434869 KZC09416.1 GL888115 EGI67172.1 GGFJ01003115 MBW52256.1 GFPF01010614 MAA21760.1 KQ981727 KYN36927.1 GGFJ01003005 MBW52146.1 GL435917 EFN72513.1 GGFM01002103 MBW22854.1 GGFJ01003077 MBW52218.1 GANP01001252 JAB83216.1 GEDV01005839 JAP82718.1 GGFJ01003026 MBW52167.1 DS234992 EEB09995.1 GACK01002810 JAA62224.1 KB632130 ERL88968.1 ADTU01012001 ADTU01012002 ADTU01012003 GGFK01006292 MBW39613.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053268
UP000007266
UP000095300
+ More
UP000094527 UP000027135 UP000092460 UP000092445 UP000092443 UP000091820 UP000078200 UP000095301 UP000235965 UP000037069 UP000218220 UP000009192 UP000053097 UP000198287 UP000015103 UP000008820 UP000279307 UP000245119 UP000075901 UP000002358 UP000075883 UP000075920 UP000192221 UP000002282 UP000092462 UP000075886 UP000000803 UP000008792 UP000007798 UP000002320 UP000008711 UP000075900 UP000000304 UP000075840 UP000075903 UP000078542 UP000007801 UP000075902 UP000215335 UP000007062 UP000001070 UP000001292 UP000075884 UP000092553 UP000075880 UP000078492 UP000268350 UP000001819 UP000019118 UP000008744 UP000036403 UP000008237 UP000075809 UP000076502 UP000007755 UP000076407 UP000078541 UP000000311 UP000009046 UP000030742 UP000005205
UP000094527 UP000027135 UP000092460 UP000092445 UP000092443 UP000091820 UP000078200 UP000095301 UP000235965 UP000037069 UP000218220 UP000009192 UP000053097 UP000198287 UP000015103 UP000008820 UP000279307 UP000245119 UP000075901 UP000002358 UP000075883 UP000075920 UP000192221 UP000002282 UP000092462 UP000075886 UP000000803 UP000008792 UP000007798 UP000002320 UP000008711 UP000075900 UP000000304 UP000075840 UP000075903 UP000078542 UP000007801 UP000075902 UP000215335 UP000007062 UP000001070 UP000001292 UP000075884 UP000092553 UP000075880 UP000078492 UP000268350 UP000001819 UP000019118 UP000008744 UP000036403 UP000008237 UP000075809 UP000076502 UP000007755 UP000076407 UP000078541 UP000000311 UP000009046 UP000030742 UP000005205
Interpro
ProteinModelPortal
H9IYX7
A0A2H1W273
A0A212ESJ3
A0A3S2TDI0
A0A194QCY9
A0A1B6DMZ0
+ More
D6WCW8 A0A1I8PIY0 A0A1D2NDW5 A0A067R8I0 A0A1B0C4G9 A0A1A9ZGE8 A0A1A9XDJ0 A0A1A9WSH0 A0A1A9VJE5 A0A1I8M3Z4 A0A2J7QZU7 D3TNS1 T1PF22 A0A0L0CQM5 A0A034VMJ3 A0A2A4J3H0 B4KBR8 A0A0K8SEJ0 A0A026VYF2 A0A226DFH0 A0A336M7L1 A0A336M7E7 A0A161M7T9 W8ARC0 A0A0P4VXM9 R4G4X2 Q16KZ1 A0A3L8D3G3 A0A1Y1M8J1 A0A1S4FX57 A0A3L8D4H0 A0A2T7PRQ9 A0A182T9B2 A0A023F8J9 K7IY66 A0A182LWN9 A0A182VYT2 A0A1W4W2D2 B4PV20 A0A1B0DCX6 A0A182Q0F1 A0A1Q3F4L1 Q9VC18 B4M648 E9ILL7 B4NI49 B0W5Z3 B3P6W1 A0A182RQ07 B4QTV0 A0A2C9GQJ3 A0A023EV17 A0A182VMZ0 A0A195CI10 B3LX08 A0A182UA66 A0A069DVW4 A0A232FCX1 A0A0V0G555 A0A0P9A1E3 Q7PXI8 B4JYA9 B4HHN0 A0A182N5G2 A0A0M4EQV3 Q6UZ77 A0A182IMK8 A0A195EMX5 A0A3B0K9U7 Q29A03 N6UNV6 B4G3K2 A0A0J7KUY0 A0A131XHL2 E2BQ40 A0A151XEX3 A0A154PBZ5 F4WFC1 A0A2M4BGS7 A0A182X9W5 A0A224Z659 A0A195F9N9 A0A2M4BGG0 E2A229 A0A2M3Z2V6 A0A2M4BHE0 V5IJL0 A0A131YVR5 A0A2M4BGK0 E0V9D9 L7MGB9 U4U9G6 A0A158NCN7 A0A2M4AFR0
D6WCW8 A0A1I8PIY0 A0A1D2NDW5 A0A067R8I0 A0A1B0C4G9 A0A1A9ZGE8 A0A1A9XDJ0 A0A1A9WSH0 A0A1A9VJE5 A0A1I8M3Z4 A0A2J7QZU7 D3TNS1 T1PF22 A0A0L0CQM5 A0A034VMJ3 A0A2A4J3H0 B4KBR8 A0A0K8SEJ0 A0A026VYF2 A0A226DFH0 A0A336M7L1 A0A336M7E7 A0A161M7T9 W8ARC0 A0A0P4VXM9 R4G4X2 Q16KZ1 A0A3L8D3G3 A0A1Y1M8J1 A0A1S4FX57 A0A3L8D4H0 A0A2T7PRQ9 A0A182T9B2 A0A023F8J9 K7IY66 A0A182LWN9 A0A182VYT2 A0A1W4W2D2 B4PV20 A0A1B0DCX6 A0A182Q0F1 A0A1Q3F4L1 Q9VC18 B4M648 E9ILL7 B4NI49 B0W5Z3 B3P6W1 A0A182RQ07 B4QTV0 A0A2C9GQJ3 A0A023EV17 A0A182VMZ0 A0A195CI10 B3LX08 A0A182UA66 A0A069DVW4 A0A232FCX1 A0A0V0G555 A0A0P9A1E3 Q7PXI8 B4JYA9 B4HHN0 A0A182N5G2 A0A0M4EQV3 Q6UZ77 A0A182IMK8 A0A195EMX5 A0A3B0K9U7 Q29A03 N6UNV6 B4G3K2 A0A0J7KUY0 A0A131XHL2 E2BQ40 A0A151XEX3 A0A154PBZ5 F4WFC1 A0A2M4BGS7 A0A182X9W5 A0A224Z659 A0A195F9N9 A0A2M4BGG0 E2A229 A0A2M3Z2V6 A0A2M4BHE0 V5IJL0 A0A131YVR5 A0A2M4BGK0 E0V9D9 L7MGB9 U4U9G6 A0A158NCN7 A0A2M4AFR0
PDB
1OZ0
E-value=0,
Score=2078
Ontologies
PATHWAY
GO
PANTHER
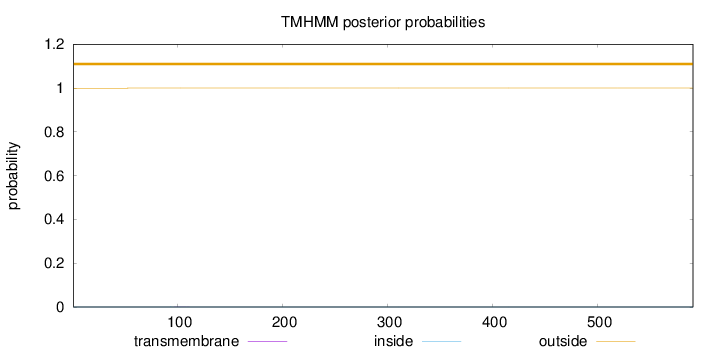
Topology
Length:
591
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00532999999999999
Exp number, first 60 AAs:
0.00174
Total prob of N-in:
0.00046
outside
1 - 591
Population Genetic Test Statistics
Pi
36.459595
Theta
69.298557
Tajima's D
-0.753244
CLR
453.337831
CSRT
0.186890655467227
Interpretation
Uncertain
