Gene
KWMTBOMO05204 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002439
Annotation
PREDICTED:_putative_methyltransferase_C9orf114_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.942
Sequence
CDS
ATGTCTGAACTAGTAGATACTAAACCTCCAGGCAAGAGTTGGCGTGAAATAAACAAAGAAAGGAAGTCACTAAAACGCAAGCGTAACGAAGAGAAAATTATCAAACGCGAGAAAAGGGTTGCCTTGGAAAAGGAAGTCCAAGATAAGGCTAAAGAAGAAGAAGAGCGCGAGAAGAAGTGTTTAGAAATTTCTACTGTTAGTATAGCTGTCCCCAGTTCTATATTAGAAAATGCTCAATCTGCAGAGCTACGTACTTATCTTGCTGGACAAATAGCCAGGGCGGCCTGTGTCTTTTGCGTCGATGAAGTAATAGTCTATGATGACATAGGTGAAAAATTAAATACAAAAAAATCTAAATTAGAGGATTCTGATGGTGTGAAAGTGGCTAGAAAGAGTTGTGTACAGTTAGCACGTATACTCCAGTACTTGGAATGTCCACAATACTTAAGAAAGCACTTTTTTCCAATACATAAAGATCTCCAGTTTGCAGGTCTACTCAATCCATTGGATGCTCCACATCATTTGAGACAGACAAATGATTTTAAATTCAGGGAAGGCATTACTATGAATAAGAAAGTGAAACCCGGTAAAGGATCGCAAGTGAATGTTGGATTGTTGCAGGATGTATCAACTGATAAATTACTAAATCCTGGTATGAGAGTTACAGTTAGAATGCTGCCTGCATTAAAAAATGGCAAGAAACTAAAAGGGAAGATTGTTAGTTTAACAACACCAAGGGCAGAGACAGGTGTTTATTGGGGTTATACTGTAAGGATTGCTAGTACATTGAGCCAAGTTTTCACTCAGTGTCCATACAAAGATGGATATGATGTTACAATAGGCACGTCTGACAGAGGATTATCAATTGACACATTACCTGATAAAGAAGTCAAATACAATCATGTTTTGATAGTTTTTGGTGGACTACATGGAATAGAAGCAGCATTGGAAAGTGATGAGCAATTAAAAGTAAATGATGCAAGTTTGTTATTCAATCATTATGTCAATGTATTACCGTCTCAAGGATCAAGGACAATTCGAACAGAGGAAGCTATATTGATAGCTTTATCAGGATTAAGAAATAAATTGCAGGCAAATAATGAACCAATGGTGTTTAAAGGTTCTGGCATTGCTCTTAGTTCTGCATTCCCTAACTTCAAACCAGATTCTGATCATTCTGGTGCAGAAAATAATGTAGATTTAAGCAGGTTTGATTAA
Protein
MSELVDTKPPGKSWREINKERKSLKRKRNEEKIIKREKRVALEKEVQDKAKEEEEREKKCLEISTVSIAVPSSILENAQSAELRTYLAGQIARAACVFCVDEVIVYDDIGEKLNTKKSKLEDSDGVKVARKSCVQLARILQYLECPQYLRKHFFPIHKDLQFAGLLNPLDAPHHLRQTNDFKFREGITMNKKVKPGKGSQVNVGLLQDVSTDKLLNPGMRVTVRMLPALKNGKKLKGKIVSLTTPRAETGVYWGYTVRIASTLSQVFTQCPYKDGYDVTIGTSDRGLSIDTLPDKEVKYNHVLIVFGGLHGIEAALESDEQLKVNDASLLFNHYVNVLPSQGSRTIRTEEAILIALSGLRNKLQANNEPMVFKGSGIALSSAFPNFKPDSDHSGAENNVDLSRFD
Summary
Uniprot
H9IYV4
A0A2H1W1T5
A0A194RNH3
A0A194QBQ2
A0A3S2NB27
S4PSF9
+ More
A0A212ESJ0 A0A0L7L7Q7 A0A2J7PDW1 D6X4M3 K7IT38 A0A067RN97 A0A232FLM4 A0A2A3E596 A0A158NZH7 F4WL90 A0A151JF60 E9J738 A0A0J7KZ85 B4GB94 A0A0M4EU77 A0A0K8TM73 Q290T9 A0A151I6K3 E2AB07 A0A182JKX2 A0A087ZXB3 B4LM14 N6U3S0 B4KU57 A0A1L8DBM9 Q8MSZ6 A0A1W4XHX2 U4U4Q9 D3PK78 A0A0L7RFL0 A0A084VEZ7 W8AHS2 A0A1B0GGS8 T1PNK2 A0A195FDD1 A0A336M442 B3MD29 A0A1I8N7Y6 A0A1J1I576 A0A1I8QEV2 E2BKZ5 A0A3B0J341 B4MQW7 A0A1B0AZM4 A0A0A1X9B0 A0A1A9YE88 B4J5A1 A0A1W4VBF7 A0A026WB06 B3N6S3 A0A310SLK3 U5EXN3 A0A1A9ZE02 A0A182HDI3 A0A1B0DB41 A0A0J9R9A3 A1Z830 Q16R76 A0A2M3ZA11 A0A2M3ZIQ1 A0A1A9WT54 B4NX31 B4QHV0 A0A1A9VIE0 A0A0K8U954 A0A2M4BP12 A0A1Y1LSB4 A0A182FI77 A0A182XZ37 A0A1Q3EY43 A0A034VX17 A0A182I3I9 A0A182UXP4 A0A182X3I1 A0A182LUV6 A0A034VVC1 A7UR97 W5JAG7 A0A182UEQ1 A0A1B6H6P7 A0A2M4AJG7 A0A182JQ37 A0A182WIN6 A0A1B6DRB7 A0A1B6JQI8 A0A182KRM8 A0A1B0G8U1 A0A1E1XF05 B0WJL6 A0A182P3Q8 A0A182NH88 A0A131YHQ1 L7M0P3
A0A212ESJ0 A0A0L7L7Q7 A0A2J7PDW1 D6X4M3 K7IT38 A0A067RN97 A0A232FLM4 A0A2A3E596 A0A158NZH7 F4WL90 A0A151JF60 E9J738 A0A0J7KZ85 B4GB94 A0A0M4EU77 A0A0K8TM73 Q290T9 A0A151I6K3 E2AB07 A0A182JKX2 A0A087ZXB3 B4LM14 N6U3S0 B4KU57 A0A1L8DBM9 Q8MSZ6 A0A1W4XHX2 U4U4Q9 D3PK78 A0A0L7RFL0 A0A084VEZ7 W8AHS2 A0A1B0GGS8 T1PNK2 A0A195FDD1 A0A336M442 B3MD29 A0A1I8N7Y6 A0A1J1I576 A0A1I8QEV2 E2BKZ5 A0A3B0J341 B4MQW7 A0A1B0AZM4 A0A0A1X9B0 A0A1A9YE88 B4J5A1 A0A1W4VBF7 A0A026WB06 B3N6S3 A0A310SLK3 U5EXN3 A0A1A9ZE02 A0A182HDI3 A0A1B0DB41 A0A0J9R9A3 A1Z830 Q16R76 A0A2M3ZA11 A0A2M3ZIQ1 A0A1A9WT54 B4NX31 B4QHV0 A0A1A9VIE0 A0A0K8U954 A0A2M4BP12 A0A1Y1LSB4 A0A182FI77 A0A182XZ37 A0A1Q3EY43 A0A034VX17 A0A182I3I9 A0A182UXP4 A0A182X3I1 A0A182LUV6 A0A034VVC1 A7UR97 W5JAG7 A0A182UEQ1 A0A1B6H6P7 A0A2M4AJG7 A0A182JQ37 A0A182WIN6 A0A1B6DRB7 A0A1B6JQI8 A0A182KRM8 A0A1B0G8U1 A0A1E1XF05 B0WJL6 A0A182P3Q8 A0A182NH88 A0A131YHQ1 L7M0P3
Pubmed
19121390
26354079
23622113
22118469
26227816
18362917
+ More
19820115 20075255 24845553 28648823 21347285 21719571 21282665 17994087 26369729 15632085 20798317 23537049 24438588 24495485 25315136 25830018 24508170 30249741 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17510324 17550304 28004739 25244985 25348373 12364791 14747013 17210077 20920257 23761445 20966253 28503490 26830274 25576852
19820115 20075255 24845553 28648823 21347285 21719571 21282665 17994087 26369729 15632085 20798317 23537049 24438588 24495485 25315136 25830018 24508170 30249741 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17510324 17550304 28004739 25244985 25348373 12364791 14747013 17210077 20920257 23761445 20966253 28503490 26830274 25576852
EMBL
BABH01010717
ODYU01005802
SOQ47055.1
KQ459984
KPJ18964.1
KQ459220
+ More
KPJ02844.1 RSAL01000293 RVE42868.1 GAIX01013858 JAA78702.1 AGBW02012778 OWR44466.1 JTDY01002386 KOB71523.1 NEVH01026136 PNF14519.1 KQ971380 EEZ97573.1 AAZX01006711 KK852570 KDR21169.1 NNAY01000061 OXU31409.1 KZ288376 PBC26664.1 ADTU01004623 GL888207 EGI65053.1 KQ979021 KYN24362.1 GL768421 EFZ11331.1 LBMM01001837 KMQ95671.1 CH479181 EDW32196.1 CP012524 ALC41200.1 GDAI01002355 JAI15248.1 CM000071 EAL25273.1 KQ978473 KYM93688.1 GL438234 EFN69283.1 CH940648 EDW59934.1 APGK01043770 KB741017 ENN75266.1 CH933808 EDW08634.1 GFDF01010350 JAV03734.1 AY118466 AAM49835.1 KB631644 ERL84950.1 BT122034 ADD31614.1 KQ414605 KOC69635.1 ATLV01012358 KE524785 KFB36541.1 GAMC01018530 JAB88025.1 AJWK01000701 KA650294 AFP64923.1 KQ981685 KYN38049.1 UFQS01000375 UFQT01000375 SSX03337.1 SSX23703.1 CH902619 EDV36344.2 CVRI01000040 CRK94906.1 GL448858 EFN83616.1 OUUW01000001 SPP73632.1 CH963849 EDW74506.1 JXJN01006375 JXJN01006376 GBXI01006771 JAD07521.1 CH916367 EDW01743.1 KK107293 QOIP01000005 EZA53287.1 RLU22489.1 CH954177 EDV58172.1 KQ760123 OAD61854.1 GANO01002465 JAB57406.1 JXUM01034236 KQ561017 KXJ80018.1 AJVK01000724 CM002911 KMY92642.1 AE013599 AAF58861.1 CH477722 EAT36903.1 GGFM01004626 MBW25377.1 GGFM01007628 MBW28379.1 CM000157 EDW89592.2 CM000362 EDX06434.1 GDHF01029127 JAI23187.1 GGFJ01005603 MBW54744.1 GEZM01052519 JAV74466.1 GFDL01014845 JAV20200.1 GAKP01012557 JAC46395.1 APCN01000213 AXCM01000276 GAKP01012558 JAC46394.1 AAAB01008807 EDO64539.1 ADMH02002032 ETN59855.1 GECU01037321 JAS70385.1 GGFK01007590 MBW40911.1 GEDC01009070 JAS28228.1 GECU01006289 JAT01418.1 CCAG010003795 CCAG010003796 GFAC01001365 JAT97823.1 DS231961 EDS29243.1 GEDV01010090 JAP78467.1 GACK01007439 JAA57595.1
KPJ02844.1 RSAL01000293 RVE42868.1 GAIX01013858 JAA78702.1 AGBW02012778 OWR44466.1 JTDY01002386 KOB71523.1 NEVH01026136 PNF14519.1 KQ971380 EEZ97573.1 AAZX01006711 KK852570 KDR21169.1 NNAY01000061 OXU31409.1 KZ288376 PBC26664.1 ADTU01004623 GL888207 EGI65053.1 KQ979021 KYN24362.1 GL768421 EFZ11331.1 LBMM01001837 KMQ95671.1 CH479181 EDW32196.1 CP012524 ALC41200.1 GDAI01002355 JAI15248.1 CM000071 EAL25273.1 KQ978473 KYM93688.1 GL438234 EFN69283.1 CH940648 EDW59934.1 APGK01043770 KB741017 ENN75266.1 CH933808 EDW08634.1 GFDF01010350 JAV03734.1 AY118466 AAM49835.1 KB631644 ERL84950.1 BT122034 ADD31614.1 KQ414605 KOC69635.1 ATLV01012358 KE524785 KFB36541.1 GAMC01018530 JAB88025.1 AJWK01000701 KA650294 AFP64923.1 KQ981685 KYN38049.1 UFQS01000375 UFQT01000375 SSX03337.1 SSX23703.1 CH902619 EDV36344.2 CVRI01000040 CRK94906.1 GL448858 EFN83616.1 OUUW01000001 SPP73632.1 CH963849 EDW74506.1 JXJN01006375 JXJN01006376 GBXI01006771 JAD07521.1 CH916367 EDW01743.1 KK107293 QOIP01000005 EZA53287.1 RLU22489.1 CH954177 EDV58172.1 KQ760123 OAD61854.1 GANO01002465 JAB57406.1 JXUM01034236 KQ561017 KXJ80018.1 AJVK01000724 CM002911 KMY92642.1 AE013599 AAF58861.1 CH477722 EAT36903.1 GGFM01004626 MBW25377.1 GGFM01007628 MBW28379.1 CM000157 EDW89592.2 CM000362 EDX06434.1 GDHF01029127 JAI23187.1 GGFJ01005603 MBW54744.1 GEZM01052519 JAV74466.1 GFDL01014845 JAV20200.1 GAKP01012557 JAC46395.1 APCN01000213 AXCM01000276 GAKP01012558 JAC46394.1 AAAB01008807 EDO64539.1 ADMH02002032 ETN59855.1 GECU01037321 JAS70385.1 GGFK01007590 MBW40911.1 GEDC01009070 JAS28228.1 GECU01006289 JAT01418.1 CCAG010003795 CCAG010003796 GFAC01001365 JAT97823.1 DS231961 EDS29243.1 GEDV01010090 JAP78467.1 GACK01007439 JAA57595.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000037510
+ More
UP000235965 UP000007266 UP000002358 UP000027135 UP000215335 UP000242457 UP000005205 UP000007755 UP000078492 UP000036403 UP000008744 UP000092553 UP000001819 UP000078542 UP000000311 UP000075880 UP000005203 UP000008792 UP000019118 UP000009192 UP000192223 UP000030742 UP000053825 UP000030765 UP000092461 UP000078541 UP000007801 UP000095301 UP000183832 UP000095300 UP000008237 UP000268350 UP000007798 UP000092460 UP000092443 UP000001070 UP000192221 UP000053097 UP000279307 UP000008711 UP000092445 UP000069940 UP000249989 UP000092462 UP000000803 UP000008820 UP000091820 UP000002282 UP000000304 UP000078200 UP000069272 UP000076408 UP000075840 UP000075903 UP000076407 UP000075883 UP000007062 UP000000673 UP000075902 UP000075881 UP000075920 UP000075882 UP000092444 UP000002320 UP000075885 UP000075884
UP000235965 UP000007266 UP000002358 UP000027135 UP000215335 UP000242457 UP000005205 UP000007755 UP000078492 UP000036403 UP000008744 UP000092553 UP000001819 UP000078542 UP000000311 UP000075880 UP000005203 UP000008792 UP000019118 UP000009192 UP000192223 UP000030742 UP000053825 UP000030765 UP000092461 UP000078541 UP000007801 UP000095301 UP000183832 UP000095300 UP000008237 UP000268350 UP000007798 UP000092460 UP000092443 UP000001070 UP000192221 UP000053097 UP000279307 UP000008711 UP000092445 UP000069940 UP000249989 UP000092462 UP000000803 UP000008820 UP000091820 UP000002282 UP000000304 UP000078200 UP000069272 UP000076408 UP000075840 UP000075903 UP000076407 UP000075883 UP000007062 UP000000673 UP000075902 UP000075881 UP000075920 UP000075882 UP000092444 UP000002320 UP000075885 UP000075884
Pfam
PF02598 Methyltrn_RNA_3
Interpro
Gene 3D
ProteinModelPortal
H9IYV4
A0A2H1W1T5
A0A194RNH3
A0A194QBQ2
A0A3S2NB27
S4PSF9
+ More
A0A212ESJ0 A0A0L7L7Q7 A0A2J7PDW1 D6X4M3 K7IT38 A0A067RN97 A0A232FLM4 A0A2A3E596 A0A158NZH7 F4WL90 A0A151JF60 E9J738 A0A0J7KZ85 B4GB94 A0A0M4EU77 A0A0K8TM73 Q290T9 A0A151I6K3 E2AB07 A0A182JKX2 A0A087ZXB3 B4LM14 N6U3S0 B4KU57 A0A1L8DBM9 Q8MSZ6 A0A1W4XHX2 U4U4Q9 D3PK78 A0A0L7RFL0 A0A084VEZ7 W8AHS2 A0A1B0GGS8 T1PNK2 A0A195FDD1 A0A336M442 B3MD29 A0A1I8N7Y6 A0A1J1I576 A0A1I8QEV2 E2BKZ5 A0A3B0J341 B4MQW7 A0A1B0AZM4 A0A0A1X9B0 A0A1A9YE88 B4J5A1 A0A1W4VBF7 A0A026WB06 B3N6S3 A0A310SLK3 U5EXN3 A0A1A9ZE02 A0A182HDI3 A0A1B0DB41 A0A0J9R9A3 A1Z830 Q16R76 A0A2M3ZA11 A0A2M3ZIQ1 A0A1A9WT54 B4NX31 B4QHV0 A0A1A9VIE0 A0A0K8U954 A0A2M4BP12 A0A1Y1LSB4 A0A182FI77 A0A182XZ37 A0A1Q3EY43 A0A034VX17 A0A182I3I9 A0A182UXP4 A0A182X3I1 A0A182LUV6 A0A034VVC1 A7UR97 W5JAG7 A0A182UEQ1 A0A1B6H6P7 A0A2M4AJG7 A0A182JQ37 A0A182WIN6 A0A1B6DRB7 A0A1B6JQI8 A0A182KRM8 A0A1B0G8U1 A0A1E1XF05 B0WJL6 A0A182P3Q8 A0A182NH88 A0A131YHQ1 L7M0P3
A0A212ESJ0 A0A0L7L7Q7 A0A2J7PDW1 D6X4M3 K7IT38 A0A067RN97 A0A232FLM4 A0A2A3E596 A0A158NZH7 F4WL90 A0A151JF60 E9J738 A0A0J7KZ85 B4GB94 A0A0M4EU77 A0A0K8TM73 Q290T9 A0A151I6K3 E2AB07 A0A182JKX2 A0A087ZXB3 B4LM14 N6U3S0 B4KU57 A0A1L8DBM9 Q8MSZ6 A0A1W4XHX2 U4U4Q9 D3PK78 A0A0L7RFL0 A0A084VEZ7 W8AHS2 A0A1B0GGS8 T1PNK2 A0A195FDD1 A0A336M442 B3MD29 A0A1I8N7Y6 A0A1J1I576 A0A1I8QEV2 E2BKZ5 A0A3B0J341 B4MQW7 A0A1B0AZM4 A0A0A1X9B0 A0A1A9YE88 B4J5A1 A0A1W4VBF7 A0A026WB06 B3N6S3 A0A310SLK3 U5EXN3 A0A1A9ZE02 A0A182HDI3 A0A1B0DB41 A0A0J9R9A3 A1Z830 Q16R76 A0A2M3ZA11 A0A2M3ZIQ1 A0A1A9WT54 B4NX31 B4QHV0 A0A1A9VIE0 A0A0K8U954 A0A2M4BP12 A0A1Y1LSB4 A0A182FI77 A0A182XZ37 A0A1Q3EY43 A0A034VX17 A0A182I3I9 A0A182UXP4 A0A182X3I1 A0A182LUV6 A0A034VVC1 A7UR97 W5JAG7 A0A182UEQ1 A0A1B6H6P7 A0A2M4AJG7 A0A182JQ37 A0A182WIN6 A0A1B6DRB7 A0A1B6JQI8 A0A182KRM8 A0A1B0G8U1 A0A1E1XF05 B0WJL6 A0A182P3Q8 A0A182NH88 A0A131YHQ1 L7M0P3
PDB
4RG1
E-value=1.41717e-95,
Score=892
Ontologies
PANTHER
Topology
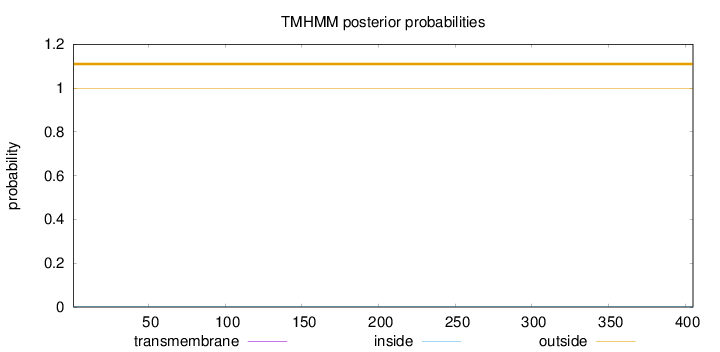
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00313
outside
1 - 405
Population Genetic Test Statistics
Pi
236.648535
Theta
200.61447
Tajima's D
0.594682
CLR
0.201044
CSRT
0.543272836358182
Interpretation
Uncertain
