Gene
KWMTBOMO05200
Pre Gene Modal
BGIBMGA002442
Annotation
neuropeptide_receptor_A19_[Bombyx_mori]
Full name
RYamide receptor
Alternative Name
Neuropeptide Y-like receptor
Location in the cell
PlasmaMembrane Reliability : 4.933
Sequence
CDS
ATGTATGAGTCGGAATCGATAGACTTTGATTCTAGTACGCTGTCAGACTTCCTTATACGCAACGTCACATCCAGTGAATACTATATAAATGCTACAGCCTCGTCCAACTTAACGAAACTAGATTACGACGCTATATGCAATCCGAGTACATCCTCAGAGAGCTTCTTTACTTCTGCGACATTCCAAACATGCGTTTATTTCATGTACTGCATCGTTTTTGTTGTGGCACTCGTTGGAAACGGACTAGTCTGCTTCGTTGTACAAACATCGCCTCGGATGAAAACGGTCACAAATTATTTCATAGTAAACCTGGCAGTGGGCGATATCCTGATGACGTTGTTCTGTGTGCCGTTTTCTTTCGTATCAATGCTTGTGCTGCGGTATTGGCCCTTTGGTGGTATTATGTGCAAAGTTGTCAACTTCTCACAAGCGGTTTCAGTGCTCGTGAGCGCATACACTTTACTAGCTATATCGATTGACCGATACATGGCAATAATGCGCCCATTGAAACCTAGGATGGGTAAAACCGCAGCAAAAATGGTTGTCGCTGGCGTGTGGGGAGGTGCTATTGCGACCGCCACCCCTATATACGTAGTATCGAAATTGGAGAGACCTGCAGAGTGGCATAAATACTGTCAACTAGATATTTGCCACGAAGAATGGGACCACGTGGAACAAAGTGAGCGTTACACCTGTGCGCTGTTGGTGCTTCAGTTTGTATTGCCACTTAGCGCTCTTGTGTGTACATATGCTCGCATTGCCCACGTAGTCTGGGGCGGACGACCACCCGGTGAAGCCGAAAGTACAAGAGATTCCAGAATGCAACGATCAAAAAGAAAGATGATCAAAATGATGGTGATTGTTGTAGCAGTATTCACGGTTTGCTGGCTCCCACTGAACATTTTTATTGCAAAAAAGGATGTCACGTTGTCAACTGACGTCAATGGACATAATCGTTTTCAGTTCAAAGCACAAATTAGCAGCTAA
Protein
MYESESIDFDSSTLSDFLIRNVTSSEYYINATASSNLTKLDYDAICNPSTSSESFFTSATFQTCVYFMYCIVFVVALVGNGLVCFVVQTSPRMKTVTNYFIVNLAVGDILMTLFCVPFSFVSMLVLRYWPFGGIMCKVVNFSQAVSVLVSAYTLLAISIDRYMAIMRPLKPRMGKTAAKMVVAGVWGGAIATATPIYVVSKLERPAEWHKYCQLDICHEEWDHVEQSERYTCALLVLQFVLPLSALVCTYARIAHVVWGGRPPGEAESTRDSRMQRSKRKMIKMMVIVVAVFTVCWLPLNIFIAKKDVTLSTDVNGHNRFQFKAQISS
Summary
Description
Receptor for the neuropeptides RYamide-1 and RYamide-2 (PubMed:21843505, PubMed:21704020). The activity of this receptor is mediated by G proteins which activate a phosphatidyl-inositol-calcium second messenger system (PubMed:21843505, PubMed:21704020). RYamide signaling may suppress feeding behavior (PubMed:21704020).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain RYamide receptor
Uniprot
B3XXN2
A0A0S1YD83
A0A2H1V2X5
A0A0L7L8U4
A0A0S1YDM6
B3XXN5
+ More
A0A194RNG8 A0A2H1VQ84 W8BKX0 W8C5P9 A0A1B6GV51 A0A1B6HNX7 A0A1B6KQU8 U3U7M9 U3U4E6 A0A1A9ZQ06 U5N1G6 B4K9P0 B4M121 A0A0R3NME0 I5ANE3 A0A0U1SQQ4 A0A1B0GFC2 A0A1A9YGR4 B3MT11 A0A1B0BK89 B4N8F0 A0A1A9UTI4 A0A1A9VZB2 A0A3B0K4J6 A0A0R3NFZ1 Q7PS45 A0A154PPP1 A0A1W4UIN3 A0A1W4U5X8 A0A2A3E4P5 A0A310SGV6 A0A087ZXB1 A0A2S2NKP8 P25931 A0A0B4LIR4 A0A2H8TU58 A0A0B4LHT6 J9JLB3 Q2QKU5 U5N1H5 A0A0L7QUK8 A0A1B6DRF1 U5N4G7 Q16Z67 B0XGN8 A0A087UNN0 A0A0P5YRX6 A0A2U9PFU7 E9G455 A0A0N8A9E0 A0A0P6G3P1 A0A147BFV4 T1KC18 A0A0P6HZ76 A0A164Z7G7
A0A194RNG8 A0A2H1VQ84 W8BKX0 W8C5P9 A0A1B6GV51 A0A1B6HNX7 A0A1B6KQU8 U3U7M9 U3U4E6 A0A1A9ZQ06 U5N1G6 B4K9P0 B4M121 A0A0R3NME0 I5ANE3 A0A0U1SQQ4 A0A1B0GFC2 A0A1A9YGR4 B3MT11 A0A1B0BK89 B4N8F0 A0A1A9UTI4 A0A1A9VZB2 A0A3B0K4J6 A0A0R3NFZ1 Q7PS45 A0A154PPP1 A0A1W4UIN3 A0A1W4U5X8 A0A2A3E4P5 A0A310SGV6 A0A087ZXB1 A0A2S2NKP8 P25931 A0A0B4LIR4 A0A2H8TU58 A0A0B4LHT6 J9JLB3 Q2QKU5 U5N1H5 A0A0L7QUK8 A0A1B6DRF1 U5N4G7 Q16Z67 B0XGN8 A0A087UNN0 A0A0P5YRX6 A0A2U9PFU7 E9G455 A0A0N8A9E0 A0A0P6G3P1 A0A147BFV4 T1KC18 A0A0P6HZ76 A0A164Z7G7
Pubmed
EMBL
AB330440
BAG68418.1
KT031017
ALM88315.1
ODYU01000259
SOQ34722.1
+ More
JTDY01002261 KOB71780.1 KT031020 ALM88318.1 AB330443 BAG68421.1 KQ459984 KPJ18959.1 ODYU01003776 SOQ42958.1 GAMC01008932 JAB97623.1 GAMC01008931 JAB97624.1 GECZ01003504 JAS66265.1 GECU01031343 JAS76363.1 GEBQ01031640 GEBQ01026161 JAT08337.1 JAT13816.1 AB817303 BAO01070.1 AB817304 BAO01071.1 KC439535 AGX85003.1 CH933806 EDW14515.1 CH940650 EDW67432.2 CM000070 KRT00006.1 EIM52478.1 JQ433061 AFJ42283.1 CCAG010006669 CH902623 EDV30401.2 KPU72827.1 JXJN01015848 CH964232 EDW81401.2 OUUW01000005 SPP80546.1 KRT00007.1 AAAB01008846 EAA06194.6 KQ435012 KZC13832.1 KZ288376 PBC26670.1 KQ760123 OAD61850.1 GGMR01005151 MBY17770.1 JN234381 AE014297 BT015303 M81490 AAT94531.1 AHN57552.1 GFXV01005684 MBW17489.1 AHN57551.1 ABLF02032190 ABLF02032192 DQ005188 AAY46405.1 KC439538 AGX85006.1 KQ414734 KOC62323.1 GEDC01021184 GEDC01009041 JAS16114.1 JAS28257.1 KC439537 AGX85005.1 CH477497 EAT39936.1 DS233047 EDS27732.1 KK120754 KFM78969.1 GDIP01053955 JAM49760.1 MG550209 AWT50643.1 GL732531 EFX85694.1 GDIP01244017 GDIP01169617 JAJ53785.1 GDIQ01049951 JAN44786.1 GEGO01005714 JAR89690.1 CAEY01001957 GDIQ01013001 JAN81736.1 LRGB01000770 KZS16035.1
JTDY01002261 KOB71780.1 KT031020 ALM88318.1 AB330443 BAG68421.1 KQ459984 KPJ18959.1 ODYU01003776 SOQ42958.1 GAMC01008932 JAB97623.1 GAMC01008931 JAB97624.1 GECZ01003504 JAS66265.1 GECU01031343 JAS76363.1 GEBQ01031640 GEBQ01026161 JAT08337.1 JAT13816.1 AB817303 BAO01070.1 AB817304 BAO01071.1 KC439535 AGX85003.1 CH933806 EDW14515.1 CH940650 EDW67432.2 CM000070 KRT00006.1 EIM52478.1 JQ433061 AFJ42283.1 CCAG010006669 CH902623 EDV30401.2 KPU72827.1 JXJN01015848 CH964232 EDW81401.2 OUUW01000005 SPP80546.1 KRT00007.1 AAAB01008846 EAA06194.6 KQ435012 KZC13832.1 KZ288376 PBC26670.1 KQ760123 OAD61850.1 GGMR01005151 MBY17770.1 JN234381 AE014297 BT015303 M81490 AAT94531.1 AHN57552.1 GFXV01005684 MBW17489.1 AHN57551.1 ABLF02032190 ABLF02032192 DQ005188 AAY46405.1 KC439538 AGX85006.1 KQ414734 KOC62323.1 GEDC01021184 GEDC01009041 JAS16114.1 JAS28257.1 KC439537 AGX85005.1 CH477497 EAT39936.1 DS233047 EDS27732.1 KK120754 KFM78969.1 GDIP01053955 JAM49760.1 MG550209 AWT50643.1 GL732531 EFX85694.1 GDIP01244017 GDIP01169617 JAJ53785.1 GDIQ01049951 JAN44786.1 GEGO01005714 JAR89690.1 CAEY01001957 GDIQ01013001 JAN81736.1 LRGB01000770 KZS16035.1
Proteomes
UP000037510
UP000053240
UP000092445
UP000009192
UP000008792
UP000001819
+ More
UP000092444 UP000092443 UP000007801 UP000092460 UP000007798 UP000078200 UP000091820 UP000268350 UP000007062 UP000076502 UP000192221 UP000242457 UP000005203 UP000000803 UP000007819 UP000053825 UP000008820 UP000002320 UP000054359 UP000000305 UP000015104 UP000076858
UP000092444 UP000092443 UP000007801 UP000092460 UP000007798 UP000078200 UP000091820 UP000268350 UP000007062 UP000076502 UP000192221 UP000242457 UP000005203 UP000000803 UP000007819 UP000053825 UP000008820 UP000002320 UP000054359 UP000000305 UP000015104 UP000076858
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
B3XXN2
A0A0S1YD83
A0A2H1V2X5
A0A0L7L8U4
A0A0S1YDM6
B3XXN5
+ More
A0A194RNG8 A0A2H1VQ84 W8BKX0 W8C5P9 A0A1B6GV51 A0A1B6HNX7 A0A1B6KQU8 U3U7M9 U3U4E6 A0A1A9ZQ06 U5N1G6 B4K9P0 B4M121 A0A0R3NME0 I5ANE3 A0A0U1SQQ4 A0A1B0GFC2 A0A1A9YGR4 B3MT11 A0A1B0BK89 B4N8F0 A0A1A9UTI4 A0A1A9VZB2 A0A3B0K4J6 A0A0R3NFZ1 Q7PS45 A0A154PPP1 A0A1W4UIN3 A0A1W4U5X8 A0A2A3E4P5 A0A310SGV6 A0A087ZXB1 A0A2S2NKP8 P25931 A0A0B4LIR4 A0A2H8TU58 A0A0B4LHT6 J9JLB3 Q2QKU5 U5N1H5 A0A0L7QUK8 A0A1B6DRF1 U5N4G7 Q16Z67 B0XGN8 A0A087UNN0 A0A0P5YRX6 A0A2U9PFU7 E9G455 A0A0N8A9E0 A0A0P6G3P1 A0A147BFV4 T1KC18 A0A0P6HZ76 A0A164Z7G7
A0A194RNG8 A0A2H1VQ84 W8BKX0 W8C5P9 A0A1B6GV51 A0A1B6HNX7 A0A1B6KQU8 U3U7M9 U3U4E6 A0A1A9ZQ06 U5N1G6 B4K9P0 B4M121 A0A0R3NME0 I5ANE3 A0A0U1SQQ4 A0A1B0GFC2 A0A1A9YGR4 B3MT11 A0A1B0BK89 B4N8F0 A0A1A9UTI4 A0A1A9VZB2 A0A3B0K4J6 A0A0R3NFZ1 Q7PS45 A0A154PPP1 A0A1W4UIN3 A0A1W4U5X8 A0A2A3E4P5 A0A310SGV6 A0A087ZXB1 A0A2S2NKP8 P25931 A0A0B4LIR4 A0A2H8TU58 A0A0B4LHT6 J9JLB3 Q2QKU5 U5N1H5 A0A0L7QUK8 A0A1B6DRF1 U5N4G7 Q16Z67 B0XGN8 A0A087UNN0 A0A0P5YRX6 A0A2U9PFU7 E9G455 A0A0N8A9E0 A0A0P6G3P1 A0A147BFV4 T1KC18 A0A0P6HZ76 A0A164Z7G7
PDB
5WS3
E-value=4.78346e-26,
Score=292
Ontologies
GO
Topology
Subcellular location
Cell membrane
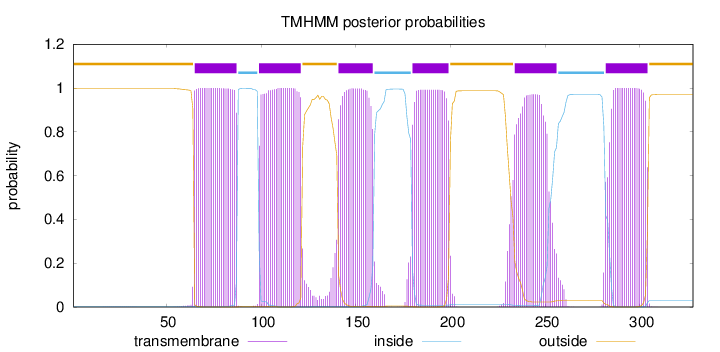
Length:
328
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
129.06733
Exp number, first 60 AAs:
0.01986
Total prob of N-in:
0.00260
outside
1 - 64
TMhelix
65 - 87
inside
88 - 98
TMhelix
99 - 121
outside
122 - 140
TMhelix
141 - 159
inside
160 - 179
TMhelix
180 - 199
outside
200 - 233
TMhelix
234 - 256
inside
257 - 281
TMhelix
282 - 304
outside
305 - 328
Population Genetic Test Statistics
Pi
269.252133
Theta
183.804369
Tajima's D
0.954896
CLR
0.519105
CSRT
0.645867706614669
Interpretation
Uncertain
