Gene
KWMTBOMO05196 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002457
Annotation
PREDICTED:_retinal_dehydrogenase_1-like_[Amyelois_transitella]
Full name
Retinal dehydrogenase 1
+ More
Aldehyde dehydrogenase, cytosolic 1
Aldehyde dehydrogenase, cytosolic 1
Alternative Name
Aldehyde dehydrogenase, cytosolic
ALDH class 1
ALDH-E1
ALHDII
Aldehyde dehydrogenase family 1 member A7
Aldehyde dehydrogenase phenobarbital-inducible
ALDH class 1
ALDH-E1
ALHDII
Aldehyde dehydrogenase family 1 member A7
Aldehyde dehydrogenase phenobarbital-inducible
Location in the cell
Cytoplasmic Reliability : 2.667
Sequence
CDS
ATGGTCCCGGAAATTAAATACACCAAGTTGTTCATTGACAACGAATGGGTTGATGCGAAAGATGGCGAAACATTTGATACATTCAGTCCACACGACGGATCAGTCATCGCTAAAGTAGCCAAAGGACAAAAGGCTGATATTGACGCCGCAGTGGCAGCGGCGAAGCGTGCGTTCCAACGTAACTCAGAATGGCGGTTGGCGGATGCCTCGCAGCGCGGAAAACTACTTTACAAGTTCGCAGATCTTATTGAACGCGACTTCGAATATCTTGCGAAACTCGAGTCATATGACAATGGCATGGTTTATAAAAATTGCCTTAGAATACTTTCAGGAATTCCTGACAGTGTAAGATACCTAGGCAGTCTGGCGGACAAAATCCAAGGCGACACAATACCAACTGATGGCGATTCATTCAGTTACACATTGAAACAACCCGTTGGAGTGTGTGGTCTCATTTTACCCTGGAACGTTCCCATCCTGATGTTTCTGAACAAGGTCGTAACTGCCTTAGCAGCAGGATGTACAGTGGTAGTGAAGCCGGCAGAACAAACTCCATTAACTGCTTTGGCATTGGCCGCACTGTTGAAAGAAGCTGGCATACCTAAGGGTGTAGTCAACATCGTGCCAGGATTCGGAGAAGATGCAGGAGTGGCGCTCACACATCACCCTGATGTTGCCAAAATTTCCTTTACAGGATCTTTGGAGGTAGGCAAACTGATCCAAAAGGCAGCTGGAACAAACAATTTGAAGCGTGTCACTCTCGAACTCGGAGGCAAGAGTCCACTAATCATAATGAATGACGCCGATTTAAATTTAGCCATCCCTTATGCTGGCAGCGGTGTATTTTTCCACCAAGGACAAATTTGCGTGGCTGCTTCCCGCTTGTACGTCCAGTCGGGTATTTACGACAAATTCGTGAAGGGGGCCGTTGAATATGCAAAAGCTATCCGGGTCGGCAATCCTTTTGACGAATCAACTGTAAACGGACCACAGATTGACGAGTCTATGATGAATAAAGTTTTGGGCTTAATAGAGACCGGAAAAAAAGAAGGAGCGAACCTACTGGCAGGTGGAGAACGTATTGGTACCGCCGGATATTATATCCAGCCTACTGTGTTTGCAGATGTTAATGATGATATGACTATTGCTAAGGAAGAGATTTTCGGTCCAGTCCAGAGCATTCTAAAGTTCGATACGTTGGAAGAAGTGATCGATCGCGCCAACAATACAAATTACGGTCTTGCAGCTGGAATCTTCACTACAAACCTTAACAACGCATTGCAGTTTAGTAAGCATGTCGAGGCCGGAACAGTTTGGGTGAATAACTACCTGACAATGGCACATCAAGCACCTTTCGGAGGATTCAAGGACTCAGGAATTGGCCGTGAAAATGGTATCAAAGCAGTTGAAGCATATTTGGAAGAGAAGACTGTCATCATTTCTCTCCCTAAAAAGCTTTGA
Protein
MVPEIKYTKLFIDNEWVDAKDGETFDTFSPHDGSVIAKVAKGQKADIDAAVAAAKRAFQRNSEWRLADASQRGKLLYKFADLIERDFEYLAKLESYDNGMVYKNCLRILSGIPDSVRYLGSLADKIQGDTIPTDGDSFSYTLKQPVGVCGLILPWNVPILMFLNKVVTALAAGCTVVVKPAEQTPLTALALAALLKEAGIPKGVVNIVPGFGEDAGVALTHHPDVAKISFTGSLEVGKLIQKAAGTNNLKRVTLELGGKSPLIIMNDADLNLAIPYAGSGVFFHQGQICVAASRLYVQSGIYDKFVKGAVEYAKAIRVGNPFDESTVNGPQIDESMMNKVLGLIETGKKEGANLLAGGERIGTAGYYIQPTVFADVNDDMTIAKEEIFGPVQSILKFDTLEEVIDRANNTNYGLAAGIFTTNLNNALQFSKHVEAGTVWVNNYLTMAHQAPFGGFKDSGIGRENGIKAVEAYLEEKTVIISLPKKL
Summary
Description
Can convert/oxidize retinaldehyde to retinoic acid. Binds free retinal and cellular retinol-binding protein-bound retinal (By similarity). May have a broader specificity and oxidize other aldehydes in vivo (By similarity).
Can oxidize benzaldehyde, propionaldehyde and acetaldehyde. No detectable activity with retinal.
Can oxidize benzaldehyde, propionaldehyde and acetaldehyde. No detectable activity with retinal.
Catalytic Activity
all-trans-retinal + H2O + NAD(+) = all-trans-retinoate + 2 H(+) + NADH
9-cis-retinal + H2O + NAD(+) = 9-cis-retinoate + 2 H(+) + NADH
11-cis-retinal + H2O + NAD(+) = 11-cis-retinoate + 2 H(+) + NADH
an aldehyde + H2O + NAD(+) = a carboxylate + 2 H(+) + NADH
9-cis-retinal + H2O + NAD(+) = 9-cis-retinoate + 2 H(+) + NADH
11-cis-retinal + H2O + NAD(+) = 11-cis-retinoate + 2 H(+) + NADH
an aldehyde + H2O + NAD(+) = a carboxylate + 2 H(+) + NADH
Biophysicochemical Properties
2.4 mM for acetaldehyde
1.6 mM for propionaldehyde
4.7 uM for benzaldehyde
1.6 mM for propionaldehyde
4.7 uM for benzaldehyde
Subunit
Homotetramer.
Similarity
Belongs to the aldehyde dehydrogenase family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Direct protein sequencing
NAD
Oxidoreductase
Phosphoprotein
Reference proteome
Feature
chain Retinal dehydrogenase 1
Uniprot
H9IYX2
A0A3S2KZ33
A0A194PUD1
A0A3S2NX34
A0A194RS96
A0A2H1VZJ0
+ More
A0A194PJZ7 A0A194RNN6 A0A2H1W5M3 A0A194PPT3 A0A2H1VS14 A0A194PP34 A0A212FDV0 A0A194RSB1 A0A0L7L392 A0A2A4J2H6 A0A2A4J196 A0A2A4JJH0 B0X5Q3 A0A3B0E4Q5 A0A1Q3FGB5 A0A182QSK4 Q16UC5 A0A182JIV1 A0A182GPP9 A0A023EVW3 A0A1S4H2G8 A0A1S4FNU3 A0A182XHT7 T1DF46 A0A2M4AEX2 A0A2M4AF40 A0A182HRD9 A0A2R5LM52 A0A182KYZ4 W5JT01 A0A182Y6E2 A0A182U026 A0A182F3L2 A0A182VIA5 A0A067QMD6 A0A084VWF2 A0A182NAW2 A0A182W8W6 A0A182PG53 A0A3S3P5E3 A0A1W7R9T2 A0A182MWM4 A0A147BAA5 Q7Q165 K7J1A8 A0A182RRC2 A0A293MT25 A0A194PHE0 A0A2H1VID4 A0A3S3NWG2 A0A212EPE8 A0A232EVC3 A0A3S3NN32 A0A0L7LH99 A0A1J1HJ07 A0A0A9WG19 A0A0K8SBS2 A0A2J7Q305 A0A2H1VRY1 A0A3L7HYV8 A0A1Z5LI79 K7FVP6 A0A3G5ANS0 A0A194R140 A0A2I0UI03 A0A0B8RTZ3 D6X3U7 A0A1D1W8T6 A0A1U7QDB4 A0A3G5AQ09 A0A2J7Q301 P86886 A0A1S3FCC6 A0A286X885 G3WEY3 A0A2J7Q307 Q2F5Y9 A0A286XZF8 T1DLI5 A0A0F7ZBD8 A0A1Y1KPZ3 A0A2H1VX41 A0A023ET21 Q6DFL9 G1KMC0 Q9PWJ3 A0A1Q3FCD3 A0A023GKS1 Q6DE83 Q16HB6 P13601 Q9YGY2 A0A286XMJ1 A0A3Q0CHQ7
A0A194PJZ7 A0A194RNN6 A0A2H1W5M3 A0A194PPT3 A0A2H1VS14 A0A194PP34 A0A212FDV0 A0A194RSB1 A0A0L7L392 A0A2A4J2H6 A0A2A4J196 A0A2A4JJH0 B0X5Q3 A0A3B0E4Q5 A0A1Q3FGB5 A0A182QSK4 Q16UC5 A0A182JIV1 A0A182GPP9 A0A023EVW3 A0A1S4H2G8 A0A1S4FNU3 A0A182XHT7 T1DF46 A0A2M4AEX2 A0A2M4AF40 A0A182HRD9 A0A2R5LM52 A0A182KYZ4 W5JT01 A0A182Y6E2 A0A182U026 A0A182F3L2 A0A182VIA5 A0A067QMD6 A0A084VWF2 A0A182NAW2 A0A182W8W6 A0A182PG53 A0A3S3P5E3 A0A1W7R9T2 A0A182MWM4 A0A147BAA5 Q7Q165 K7J1A8 A0A182RRC2 A0A293MT25 A0A194PHE0 A0A2H1VID4 A0A3S3NWG2 A0A212EPE8 A0A232EVC3 A0A3S3NN32 A0A0L7LH99 A0A1J1HJ07 A0A0A9WG19 A0A0K8SBS2 A0A2J7Q305 A0A2H1VRY1 A0A3L7HYV8 A0A1Z5LI79 K7FVP6 A0A3G5ANS0 A0A194R140 A0A2I0UI03 A0A0B8RTZ3 D6X3U7 A0A1D1W8T6 A0A1U7QDB4 A0A3G5AQ09 A0A2J7Q301 P86886 A0A1S3FCC6 A0A286X885 G3WEY3 A0A2J7Q307 Q2F5Y9 A0A286XZF8 T1DLI5 A0A0F7ZBD8 A0A1Y1KPZ3 A0A2H1VX41 A0A023ET21 Q6DFL9 G1KMC0 Q9PWJ3 A0A1Q3FCD3 A0A023GKS1 Q6DE83 Q16HB6 P13601 Q9YGY2 A0A286XMJ1 A0A3Q0CHQ7
EC Number
1.2.1.-
1.2.1.36
1.2.1.3
1.2.1.36
1.2.1.3
Pubmed
19121390
26354079
22118469
26227816
17510324
26483478
+ More
24945155 12364791 20966253 20920257 23761445 25244985 24845553 24438588 20075255 28648823 25401762 29704459 28528879 17381049 25476704 18362917 19820115 27649274 21993624 21709235 23758969 28004739 10085078 2753900 10998257 10091603 28071753
24945155 12364791 20966253 20920257 23761445 25244985 24845553 24438588 20075255 28648823 25401762 29704459 28528879 17381049 25476704 18362917 19820115 27649274 21993624 21709235 23758969 28004739 10085078 2753900 10998257 10091603 28071753
EMBL
BABH01010692
BABH01010693
BABH01010694
BABH01010695
BABH01010696
RSAL01000402
+ More
RVE41911.1 KQ459597 KPI94745.1 RSAL01000037 RVE51196.1 KQ459984 KPJ18941.1 ODYU01005422 SOQ46251.1 KQ459606 KPI91415.1 KPJ18940.1 ODYU01006360 SOQ48142.1 KPI94744.1 ODYU01004072 SOQ43566.1 KPI94743.1 AGBW02008996 OWR51929.1 KPJ18956.1 JTDY01003290 KOB69791.1 NWSH01003948 PCG65640.1 PCG65639.1 NWSH01001358 PCG71550.1 DS232392 EDS41034.1 RBVL01000220 RKO10496.1 GFDL01008464 JAV26581.1 AXCN02000654 CH477625 EAT38124.1 JXUM01078869 KQ563093 KXJ74487.1 GAPW01001149 JAC12449.1 AAAB01008980 GAMD01003251 JAA98339.1 GGFK01006020 MBW39341.1 GGFK01006088 MBW39409.1 APCN01001694 GGLE01006443 MBY10569.1 ADMH02000539 ETN66060.1 KK853170 KDR10336.1 ATLV01017583 KE525174 KFB42296.1 NCKU01001310 RWS12467.1 GFAH01000487 JAV47902.1 AXCM01004965 GEIB01000036 JAR87698.1 EAA14068.3 GFWV01019295 MAA44023.1 KQ459603 KPI92727.1 ODYU01002733 SOQ40599.1 NCKU01002086 RWS10468.1 AGBW02013494 OWR43337.1 NNAY01002022 OXU22287.1 NCKU01007299 RWS02740.1 JTDY01001120 KOB74817.1 CVRI01000006 CRK88023.1 GBHO01036212 JAG07392.1 GBRD01015069 JAG50757.1 NEVH01019075 PNF22959.1 ODYU01004073 SOQ43567.1 RAZU01000133 RLQ71138.1 GFJQ02000226 JAW06744.1 AGCU01194510 AGCU01194511 AGCU01194512 AGCU01194513 AGCU01194514 AGCU01194515 AGCU01194516 AGCU01194517 AGCU01194518 MH990439 AYV88986.1 KQ460883 KPJ11229.1 KZ505748 PKU45677.1 GBSH01002235 JAG66791.1 KQ971326 EEZ97369.1 BDGG01000018 GAV08728.1 MH979760 AYV89215.1 PNF22957.1 AAKN02026169 AAKN02026170 AAKN02026171 AAKN02026172 AAKN02026173 AEFK01077107 AEFK01077108 AEFK01077109 AEFK01077110 AEFK01077111 AEFK01077112 AEFK01077113 AEFK01077114 AEFK01077115 AEFK01077116 PNF22956.1 BABH01005570 BABH01005571 BABH01005572 BABH01005573 DQ311283 ABD36228.1 GAAZ01002032 JAA95911.1 GBEX01002791 JAI11769.1 GEZM01076997 JAV63479.1 ODYU01004657 SOQ44774.1 GAPW01001148 JAC12450.1 BC076716 AAH76716.1 AB016718 BAA76412.1 GFDL01009835 JAV25210.1 GBBM01000946 JAC34472.1 BC077256 AAH77256.1 CH478186 EAT33638.1 M23995 AF061833 AAC69552.1
RVE41911.1 KQ459597 KPI94745.1 RSAL01000037 RVE51196.1 KQ459984 KPJ18941.1 ODYU01005422 SOQ46251.1 KQ459606 KPI91415.1 KPJ18940.1 ODYU01006360 SOQ48142.1 KPI94744.1 ODYU01004072 SOQ43566.1 KPI94743.1 AGBW02008996 OWR51929.1 KPJ18956.1 JTDY01003290 KOB69791.1 NWSH01003948 PCG65640.1 PCG65639.1 NWSH01001358 PCG71550.1 DS232392 EDS41034.1 RBVL01000220 RKO10496.1 GFDL01008464 JAV26581.1 AXCN02000654 CH477625 EAT38124.1 JXUM01078869 KQ563093 KXJ74487.1 GAPW01001149 JAC12449.1 AAAB01008980 GAMD01003251 JAA98339.1 GGFK01006020 MBW39341.1 GGFK01006088 MBW39409.1 APCN01001694 GGLE01006443 MBY10569.1 ADMH02000539 ETN66060.1 KK853170 KDR10336.1 ATLV01017583 KE525174 KFB42296.1 NCKU01001310 RWS12467.1 GFAH01000487 JAV47902.1 AXCM01004965 GEIB01000036 JAR87698.1 EAA14068.3 GFWV01019295 MAA44023.1 KQ459603 KPI92727.1 ODYU01002733 SOQ40599.1 NCKU01002086 RWS10468.1 AGBW02013494 OWR43337.1 NNAY01002022 OXU22287.1 NCKU01007299 RWS02740.1 JTDY01001120 KOB74817.1 CVRI01000006 CRK88023.1 GBHO01036212 JAG07392.1 GBRD01015069 JAG50757.1 NEVH01019075 PNF22959.1 ODYU01004073 SOQ43567.1 RAZU01000133 RLQ71138.1 GFJQ02000226 JAW06744.1 AGCU01194510 AGCU01194511 AGCU01194512 AGCU01194513 AGCU01194514 AGCU01194515 AGCU01194516 AGCU01194517 AGCU01194518 MH990439 AYV88986.1 KQ460883 KPJ11229.1 KZ505748 PKU45677.1 GBSH01002235 JAG66791.1 KQ971326 EEZ97369.1 BDGG01000018 GAV08728.1 MH979760 AYV89215.1 PNF22957.1 AAKN02026169 AAKN02026170 AAKN02026171 AAKN02026172 AAKN02026173 AEFK01077107 AEFK01077108 AEFK01077109 AEFK01077110 AEFK01077111 AEFK01077112 AEFK01077113 AEFK01077114 AEFK01077115 AEFK01077116 PNF22956.1 BABH01005570 BABH01005571 BABH01005572 BABH01005573 DQ311283 ABD36228.1 GAAZ01002032 JAA95911.1 GBEX01002791 JAI11769.1 GEZM01076997 JAV63479.1 ODYU01004657 SOQ44774.1 GAPW01001148 JAC12450.1 BC076716 AAH76716.1 AB016718 BAA76412.1 GFDL01009835 JAV25210.1 GBBM01000946 JAC34472.1 BC077256 AAH77256.1 CH478186 EAT33638.1 M23995 AF061833 AAC69552.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000218220 UP000002320 UP000276569 UP000075886 UP000008820 UP000075880 UP000069940 UP000249989 UP000076407 UP000075840 UP000075882 UP000000673 UP000076408 UP000075902 UP000069272 UP000075903 UP000027135 UP000030765 UP000075884 UP000075920 UP000075885 UP000285301 UP000075883 UP000007062 UP000002358 UP000075900 UP000215335 UP000183832 UP000235965 UP000273346 UP000007267 UP000007266 UP000186922 UP000189706 UP000081671 UP000005447 UP000007648 UP000001646 UP000002494
UP000218220 UP000002320 UP000276569 UP000075886 UP000008820 UP000075880 UP000069940 UP000249989 UP000076407 UP000075840 UP000075882 UP000000673 UP000076408 UP000075902 UP000069272 UP000075903 UP000027135 UP000030765 UP000075884 UP000075920 UP000075885 UP000285301 UP000075883 UP000007062 UP000002358 UP000075900 UP000215335 UP000183832 UP000235965 UP000273346 UP000007267 UP000007266 UP000186922 UP000189706 UP000081671 UP000005447 UP000007648 UP000001646 UP000002494
Pfam
PF00171 Aldedh
Interpro
SUPFAM
SSF53720
SSF53720
Gene 3D
ProteinModelPortal
H9IYX2
A0A3S2KZ33
A0A194PUD1
A0A3S2NX34
A0A194RS96
A0A2H1VZJ0
+ More
A0A194PJZ7 A0A194RNN6 A0A2H1W5M3 A0A194PPT3 A0A2H1VS14 A0A194PP34 A0A212FDV0 A0A194RSB1 A0A0L7L392 A0A2A4J2H6 A0A2A4J196 A0A2A4JJH0 B0X5Q3 A0A3B0E4Q5 A0A1Q3FGB5 A0A182QSK4 Q16UC5 A0A182JIV1 A0A182GPP9 A0A023EVW3 A0A1S4H2G8 A0A1S4FNU3 A0A182XHT7 T1DF46 A0A2M4AEX2 A0A2M4AF40 A0A182HRD9 A0A2R5LM52 A0A182KYZ4 W5JT01 A0A182Y6E2 A0A182U026 A0A182F3L2 A0A182VIA5 A0A067QMD6 A0A084VWF2 A0A182NAW2 A0A182W8W6 A0A182PG53 A0A3S3P5E3 A0A1W7R9T2 A0A182MWM4 A0A147BAA5 Q7Q165 K7J1A8 A0A182RRC2 A0A293MT25 A0A194PHE0 A0A2H1VID4 A0A3S3NWG2 A0A212EPE8 A0A232EVC3 A0A3S3NN32 A0A0L7LH99 A0A1J1HJ07 A0A0A9WG19 A0A0K8SBS2 A0A2J7Q305 A0A2H1VRY1 A0A3L7HYV8 A0A1Z5LI79 K7FVP6 A0A3G5ANS0 A0A194R140 A0A2I0UI03 A0A0B8RTZ3 D6X3U7 A0A1D1W8T6 A0A1U7QDB4 A0A3G5AQ09 A0A2J7Q301 P86886 A0A1S3FCC6 A0A286X885 G3WEY3 A0A2J7Q307 Q2F5Y9 A0A286XZF8 T1DLI5 A0A0F7ZBD8 A0A1Y1KPZ3 A0A2H1VX41 A0A023ET21 Q6DFL9 G1KMC0 Q9PWJ3 A0A1Q3FCD3 A0A023GKS1 Q6DE83 Q16HB6 P13601 Q9YGY2 A0A286XMJ1 A0A3Q0CHQ7
A0A194PJZ7 A0A194RNN6 A0A2H1W5M3 A0A194PPT3 A0A2H1VS14 A0A194PP34 A0A212FDV0 A0A194RSB1 A0A0L7L392 A0A2A4J2H6 A0A2A4J196 A0A2A4JJH0 B0X5Q3 A0A3B0E4Q5 A0A1Q3FGB5 A0A182QSK4 Q16UC5 A0A182JIV1 A0A182GPP9 A0A023EVW3 A0A1S4H2G8 A0A1S4FNU3 A0A182XHT7 T1DF46 A0A2M4AEX2 A0A2M4AF40 A0A182HRD9 A0A2R5LM52 A0A182KYZ4 W5JT01 A0A182Y6E2 A0A182U026 A0A182F3L2 A0A182VIA5 A0A067QMD6 A0A084VWF2 A0A182NAW2 A0A182W8W6 A0A182PG53 A0A3S3P5E3 A0A1W7R9T2 A0A182MWM4 A0A147BAA5 Q7Q165 K7J1A8 A0A182RRC2 A0A293MT25 A0A194PHE0 A0A2H1VID4 A0A3S3NWG2 A0A212EPE8 A0A232EVC3 A0A3S3NN32 A0A0L7LH99 A0A1J1HJ07 A0A0A9WG19 A0A0K8SBS2 A0A2J7Q305 A0A2H1VRY1 A0A3L7HYV8 A0A1Z5LI79 K7FVP6 A0A3G5ANS0 A0A194R140 A0A2I0UI03 A0A0B8RTZ3 D6X3U7 A0A1D1W8T6 A0A1U7QDB4 A0A3G5AQ09 A0A2J7Q301 P86886 A0A1S3FCC6 A0A286X885 G3WEY3 A0A2J7Q307 Q2F5Y9 A0A286XZF8 T1DLI5 A0A0F7ZBD8 A0A1Y1KPZ3 A0A2H1VX41 A0A023ET21 Q6DFL9 G1KMC0 Q9PWJ3 A0A1Q3FCD3 A0A023GKS1 Q6DE83 Q16HB6 P13601 Q9YGY2 A0A286XMJ1 A0A3Q0CHQ7
PDB
5L2O
E-value=7.28088e-148,
Score=1344
Ontologies
KEGG
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Cytoplasm
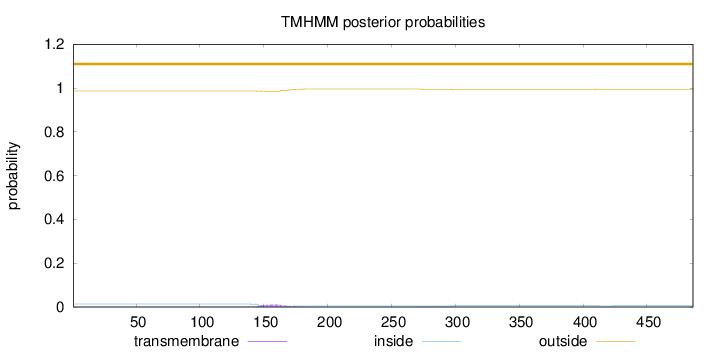
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26137
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01364
outside
1 - 486
Population Genetic Test Statistics
Pi
171.746592
Theta
143.64057
Tajima's D
0.586666
CLR
0
CSRT
0.538823058847058
Interpretation
Uncertain
