Gene
KWMTBOMO05192
Pre Gene Modal
BGIBMGA002454
Annotation
PREDICTED:_complex_III_assembly_factor_LYRM7_[Papilio_machaon]
Full name
Palmitoyltransferase
Location in the cell
Nuclear Reliability : 1.988
Sequence
CDS
ATGATGACAAGACGTTTGGTTTTACAAAGCTTCAAGAAATTGCACAGAACTAGAATGAAAGTCTTCGAAGGCGACGAACGTGCTTTATTAGCGGCACGAATAAAAATAAATGAAGAATTTCGCAAAAACAAAAATGTTCAAAATGAAAACTCGATTAAAGCGATGATTGCATTTGGAGAGGATATAGAAAGGGAATTAAGGACTCAAGTGATTCAAGCCCGAGAAATAAAACCAGGTGTTTATGAAGCAAGAATTACTGATGACACTGTGAAATTAGACAACATACCTTTCAATGATCATGCTATATTAGAAGATGGGACAGAAGCACCACGACCATGCTGTCAAGATAAAACCAAATAA
Protein
MMTRRLVLQSFKKLHRTRMKVFEGDERALLAARIKINEEFRKNKNVQNENSIKAMIAFGEDIERELRTQVIQAREIKPGVYEARITDDTVKLDNIPFNDHAILEDGTEAPRPCCQDKTK
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the complex I LYR family.
Belongs to the DHHC palmitoyltransferase family.
Belongs to the DHHC palmitoyltransferase family.
Uniprot
A0A2H1VNE0
S4NSR9
A0A194PPS8
A0A194RN62
A0A1E1WSD8
A0A212FDZ1
+ More
A0A0A9YGB8 A0A1B6JNY1 A0A1B6FYF8 A0A1B6KV98 N6U2S7 A0A1Y1KLQ4 W8C6A6 A0A087ZRJ4 B4PV41 B4K5I5 A2VEP3 A0A1W4VQ80 B4QTS8 Q0KI15 A2VES4 A0A0A1X235 A0A2A3E005 B4HHK7 A0A1B6C4T0 B3P6Y4 A0A0K8WHP6 A0A0K8VI95 A0A0N7Z9A3 A0A034V8X6 T1HEJ5 B3LYA7 A2VES5 A0A1B0BTE6 A0A0M5J583 A0A2P8Z1A0 A0A067RW51 A0A232F8N1 A0A1A9XK00 A0A1A9UQW5 A2VEU5 A0A3B0K8B6 B4LWD9 A0A336M008 A0A1A9ZWS4 A0A1B6KZT5 B4NGP7 A0A1B0G2Z3 A0A1S3CTT7 A0A1A9X1P1 A0A1B6FGY2 K7JKY9 D2A462 B4GPD3 U5EX01 A0A1I8QE30 A0A0R3NEZ4 B5DXS5 A0A1I8N1D4 A0A0L0CCK9 A0A0L8FIU1 A0A0L7RKU9 A0A1W4WV46 E9IID6 A0A0K8TPC6 A0A0J7NJS0 A0A3Q3N7J3 A0A3Q3Q099 A0A151WIH9 A0A3Q3A0E7 A0A3Q3GAZ1 A0A0P4WD13 A0A3Q3N7I8 A0A1A7W802 A0A195FV07 F4WMW2 A0A182S4J6 A0A182PIQ0 A0A0T6AWG4 A0A2M3ZDL7 A0A2I4D691 A0A182WBC2 A0A026X3S4 A0A3B4AGA4 A0A182L3U7 A0A2M4CVR9 A0A2M4C3T6 A0A182UUT3 A0A182K2A4 A0A182TI61 A0A182WWB8 Q7PTD0 A0A182I6J8 A0A2M4AYB1 A0A182FGS3 A0A3P9M6R6 A0A3B3I281 A0A2M4C3S6
A0A0A9YGB8 A0A1B6JNY1 A0A1B6FYF8 A0A1B6KV98 N6U2S7 A0A1Y1KLQ4 W8C6A6 A0A087ZRJ4 B4PV41 B4K5I5 A2VEP3 A0A1W4VQ80 B4QTS8 Q0KI15 A2VES4 A0A0A1X235 A0A2A3E005 B4HHK7 A0A1B6C4T0 B3P6Y4 A0A0K8WHP6 A0A0K8VI95 A0A0N7Z9A3 A0A034V8X6 T1HEJ5 B3LYA7 A2VES5 A0A1B0BTE6 A0A0M5J583 A0A2P8Z1A0 A0A067RW51 A0A232F8N1 A0A1A9XK00 A0A1A9UQW5 A2VEU5 A0A3B0K8B6 B4LWD9 A0A336M008 A0A1A9ZWS4 A0A1B6KZT5 B4NGP7 A0A1B0G2Z3 A0A1S3CTT7 A0A1A9X1P1 A0A1B6FGY2 K7JKY9 D2A462 B4GPD3 U5EX01 A0A1I8QE30 A0A0R3NEZ4 B5DXS5 A0A1I8N1D4 A0A0L0CCK9 A0A0L8FIU1 A0A0L7RKU9 A0A1W4WV46 E9IID6 A0A0K8TPC6 A0A0J7NJS0 A0A3Q3N7J3 A0A3Q3Q099 A0A151WIH9 A0A3Q3A0E7 A0A3Q3GAZ1 A0A0P4WD13 A0A3Q3N7I8 A0A1A7W802 A0A195FV07 F4WMW2 A0A182S4J6 A0A182PIQ0 A0A0T6AWG4 A0A2M3ZDL7 A0A2I4D691 A0A182WBC2 A0A026X3S4 A0A3B4AGA4 A0A182L3U7 A0A2M4CVR9 A0A2M4C3T6 A0A182UUT3 A0A182K2A4 A0A182TI61 A0A182WWB8 Q7PTD0 A0A182I6J8 A0A2M4AYB1 A0A182FGS3 A0A3P9M6R6 A0A3B3I281 A0A2M4C3S6
EC Number
2.3.1.225
Pubmed
23622113
26354079
22118469
25401762
23537049
28004739
+ More
24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 27129103 25348373 29403074 24845553 28648823 20075255 18362917 19820115 15632085 25315136 26108605 21282665 26369729 21719571 24508170 25463417 20966253 12364791 14747013 17210077 17554307
24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 27129103 25348373 29403074 24845553 28648823 20075255 18362917 19820115 15632085 25315136 26108605 21282665 26369729 21719571 24508170 25463417 20966253 12364791 14747013 17210077 17554307
EMBL
ODYU01003479
SOQ42333.1
GAIX01012456
JAA80104.1
KQ459597
KPI94739.1
+ More
KQ459984 KPJ18952.1 GDQN01003566 GDQN01001091 JAT87488.1 JAT89963.1 AGBW02008996 OWR51933.1 GBHO01011512 GBRD01005593 JAG32092.1 JAG60228.1 GECU01006796 GECU01006283 GECU01002195 JAT00911.1 JAT01424.1 JAT05512.1 GECZ01014553 JAS55216.1 GEBQ01024617 JAT15360.1 APGK01051789 KB741207 KB632426 ENN72872.1 ERL95381.1 GEZM01081798 JAV61528.1 GAMC01004394 GAMC01004392 JAC02162.1 CM000160 EDW98890.1 CH933806 EDW14022.1 BT030212 ABN49351.1 CM000364 EDX14282.1 AE014297 KX531456 ABI31206.2 ANY27266.1 BT030243 ABN49382.1 GBXI01009564 JAD04728.1 KZ288510 PBC25123.1 CH480815 EDW43549.1 GEDC01028904 JAS08394.1 CH954182 EDV53804.1 GDHF01001641 JAI50673.1 GDHF01028786 GDHF01022015 GDHF01013698 JAI23528.1 JAI30299.1 JAI38616.1 GDKW01001107 JAI55488.1 GAKP01020378 GAKP01020376 GAKP01020375 JAC38577.1 ACPB03010483 CH902617 EDV44011.1 BT030244 ABN49383.1 JXJN01020100 CP012526 ALC47211.1 PYGN01000245 PSN50273.1 KK852427 KDR24099.1 NNAY01000716 OXU26828.1 BT030264 ABN49403.1 OUUW01000005 SPP81261.1 CH940650 EDW66576.2 UFQT01000215 SSX21817.1 GEBQ01023072 JAT16905.1 CH964272 EDW84394.1 CCAG010003320 GECZ01020301 JAS49468.1 KQ971348 KQ971346 EFA04839.1 EFA05482.1 CH479186 EDW39016.1 GANO01002746 JAB57125.1 CM000070 KRS99594.1 EDY67516.2 JRES01000608 KNC29947.1 KQ430665 KOF63828.1 KQ414569 KOC71361.1 GL763459 EFZ19686.1 GDAI01001399 JAI16204.1 LBMM01004174 KMQ92735.1 KQ983089 KYQ47634.1 GDRN01049465 JAI66553.1 HADW01000430 HADX01013851 SBP01830.1 KQ981261 KYN44122.1 GL888223 EGI64432.1 LJIG01022681 KRT79275.1 GGFM01005896 MBW26647.1 KK107019 EZA62631.1 GGFL01004750 MBW68928.1 GGFJ01010846 MBW59987.1 AAAB01008807 EAA04084.2 APCN01003655 GGFK01012414 MBW45735.1 GGFJ01010845 MBW59986.1
KQ459984 KPJ18952.1 GDQN01003566 GDQN01001091 JAT87488.1 JAT89963.1 AGBW02008996 OWR51933.1 GBHO01011512 GBRD01005593 JAG32092.1 JAG60228.1 GECU01006796 GECU01006283 GECU01002195 JAT00911.1 JAT01424.1 JAT05512.1 GECZ01014553 JAS55216.1 GEBQ01024617 JAT15360.1 APGK01051789 KB741207 KB632426 ENN72872.1 ERL95381.1 GEZM01081798 JAV61528.1 GAMC01004394 GAMC01004392 JAC02162.1 CM000160 EDW98890.1 CH933806 EDW14022.1 BT030212 ABN49351.1 CM000364 EDX14282.1 AE014297 KX531456 ABI31206.2 ANY27266.1 BT030243 ABN49382.1 GBXI01009564 JAD04728.1 KZ288510 PBC25123.1 CH480815 EDW43549.1 GEDC01028904 JAS08394.1 CH954182 EDV53804.1 GDHF01001641 JAI50673.1 GDHF01028786 GDHF01022015 GDHF01013698 JAI23528.1 JAI30299.1 JAI38616.1 GDKW01001107 JAI55488.1 GAKP01020378 GAKP01020376 GAKP01020375 JAC38577.1 ACPB03010483 CH902617 EDV44011.1 BT030244 ABN49383.1 JXJN01020100 CP012526 ALC47211.1 PYGN01000245 PSN50273.1 KK852427 KDR24099.1 NNAY01000716 OXU26828.1 BT030264 ABN49403.1 OUUW01000005 SPP81261.1 CH940650 EDW66576.2 UFQT01000215 SSX21817.1 GEBQ01023072 JAT16905.1 CH964272 EDW84394.1 CCAG010003320 GECZ01020301 JAS49468.1 KQ971348 KQ971346 EFA04839.1 EFA05482.1 CH479186 EDW39016.1 GANO01002746 JAB57125.1 CM000070 KRS99594.1 EDY67516.2 JRES01000608 KNC29947.1 KQ430665 KOF63828.1 KQ414569 KOC71361.1 GL763459 EFZ19686.1 GDAI01001399 JAI16204.1 LBMM01004174 KMQ92735.1 KQ983089 KYQ47634.1 GDRN01049465 JAI66553.1 HADW01000430 HADX01013851 SBP01830.1 KQ981261 KYN44122.1 GL888223 EGI64432.1 LJIG01022681 KRT79275.1 GGFM01005896 MBW26647.1 KK107019 EZA62631.1 GGFL01004750 MBW68928.1 GGFJ01010846 MBW59987.1 AAAB01008807 EAA04084.2 APCN01003655 GGFK01012414 MBW45735.1 GGFJ01010845 MBW59986.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000019118
UP000030742
UP000005203
+ More
UP000002282 UP000009192 UP000192221 UP000000304 UP000000803 UP000242457 UP000001292 UP000008711 UP000015103 UP000007801 UP000092460 UP000092553 UP000245037 UP000027135 UP000215335 UP000092443 UP000078200 UP000268350 UP000008792 UP000092445 UP000007798 UP000092444 UP000079169 UP000091820 UP000002358 UP000007266 UP000008744 UP000095300 UP000001819 UP000095301 UP000037069 UP000053454 UP000053825 UP000192223 UP000036403 UP000261640 UP000261600 UP000075809 UP000264800 UP000261660 UP000078541 UP000007755 UP000075900 UP000075885 UP000192220 UP000075920 UP000053097 UP000261520 UP000075882 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000265180 UP000001038
UP000002282 UP000009192 UP000192221 UP000000304 UP000000803 UP000242457 UP000001292 UP000008711 UP000015103 UP000007801 UP000092460 UP000092553 UP000245037 UP000027135 UP000215335 UP000092443 UP000078200 UP000268350 UP000008792 UP000092445 UP000007798 UP000092444 UP000079169 UP000091820 UP000002358 UP000007266 UP000008744 UP000095300 UP000001819 UP000095301 UP000037069 UP000053454 UP000053825 UP000192223 UP000036403 UP000261640 UP000261600 UP000075809 UP000264800 UP000261660 UP000078541 UP000007755 UP000075900 UP000075885 UP000192220 UP000075920 UP000053097 UP000261520 UP000075882 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000265180 UP000001038
PRIDE
ProteinModelPortal
A0A2H1VNE0
S4NSR9
A0A194PPS8
A0A194RN62
A0A1E1WSD8
A0A212FDZ1
+ More
A0A0A9YGB8 A0A1B6JNY1 A0A1B6FYF8 A0A1B6KV98 N6U2S7 A0A1Y1KLQ4 W8C6A6 A0A087ZRJ4 B4PV41 B4K5I5 A2VEP3 A0A1W4VQ80 B4QTS8 Q0KI15 A2VES4 A0A0A1X235 A0A2A3E005 B4HHK7 A0A1B6C4T0 B3P6Y4 A0A0K8WHP6 A0A0K8VI95 A0A0N7Z9A3 A0A034V8X6 T1HEJ5 B3LYA7 A2VES5 A0A1B0BTE6 A0A0M5J583 A0A2P8Z1A0 A0A067RW51 A0A232F8N1 A0A1A9XK00 A0A1A9UQW5 A2VEU5 A0A3B0K8B6 B4LWD9 A0A336M008 A0A1A9ZWS4 A0A1B6KZT5 B4NGP7 A0A1B0G2Z3 A0A1S3CTT7 A0A1A9X1P1 A0A1B6FGY2 K7JKY9 D2A462 B4GPD3 U5EX01 A0A1I8QE30 A0A0R3NEZ4 B5DXS5 A0A1I8N1D4 A0A0L0CCK9 A0A0L8FIU1 A0A0L7RKU9 A0A1W4WV46 E9IID6 A0A0K8TPC6 A0A0J7NJS0 A0A3Q3N7J3 A0A3Q3Q099 A0A151WIH9 A0A3Q3A0E7 A0A3Q3GAZ1 A0A0P4WD13 A0A3Q3N7I8 A0A1A7W802 A0A195FV07 F4WMW2 A0A182S4J6 A0A182PIQ0 A0A0T6AWG4 A0A2M3ZDL7 A0A2I4D691 A0A182WBC2 A0A026X3S4 A0A3B4AGA4 A0A182L3U7 A0A2M4CVR9 A0A2M4C3T6 A0A182UUT3 A0A182K2A4 A0A182TI61 A0A182WWB8 Q7PTD0 A0A182I6J8 A0A2M4AYB1 A0A182FGS3 A0A3P9M6R6 A0A3B3I281 A0A2M4C3S6
A0A0A9YGB8 A0A1B6JNY1 A0A1B6FYF8 A0A1B6KV98 N6U2S7 A0A1Y1KLQ4 W8C6A6 A0A087ZRJ4 B4PV41 B4K5I5 A2VEP3 A0A1W4VQ80 B4QTS8 Q0KI15 A2VES4 A0A0A1X235 A0A2A3E005 B4HHK7 A0A1B6C4T0 B3P6Y4 A0A0K8WHP6 A0A0K8VI95 A0A0N7Z9A3 A0A034V8X6 T1HEJ5 B3LYA7 A2VES5 A0A1B0BTE6 A0A0M5J583 A0A2P8Z1A0 A0A067RW51 A0A232F8N1 A0A1A9XK00 A0A1A9UQW5 A2VEU5 A0A3B0K8B6 B4LWD9 A0A336M008 A0A1A9ZWS4 A0A1B6KZT5 B4NGP7 A0A1B0G2Z3 A0A1S3CTT7 A0A1A9X1P1 A0A1B6FGY2 K7JKY9 D2A462 B4GPD3 U5EX01 A0A1I8QE30 A0A0R3NEZ4 B5DXS5 A0A1I8N1D4 A0A0L0CCK9 A0A0L8FIU1 A0A0L7RKU9 A0A1W4WV46 E9IID6 A0A0K8TPC6 A0A0J7NJS0 A0A3Q3N7J3 A0A3Q3Q099 A0A151WIH9 A0A3Q3A0E7 A0A3Q3GAZ1 A0A0P4WD13 A0A3Q3N7I8 A0A1A7W802 A0A195FV07 F4WMW2 A0A182S4J6 A0A182PIQ0 A0A0T6AWG4 A0A2M3ZDL7 A0A2I4D691 A0A182WBC2 A0A026X3S4 A0A3B4AGA4 A0A182L3U7 A0A2M4CVR9 A0A2M4C3T6 A0A182UUT3 A0A182K2A4 A0A182TI61 A0A182WWB8 Q7PTD0 A0A182I6J8 A0A2M4AYB1 A0A182FGS3 A0A3P9M6R6 A0A3B3I281 A0A2M4C3S6
Ontologies
KEGG
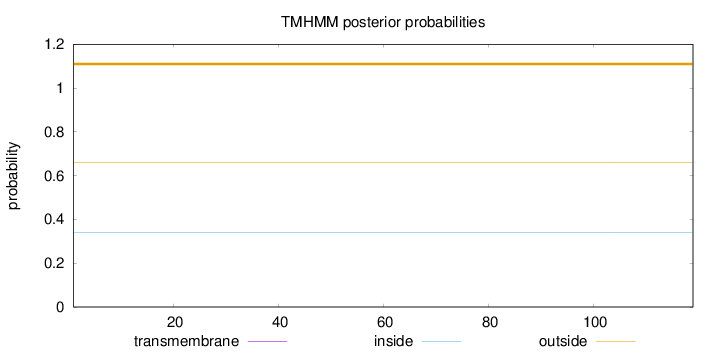
Topology
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.33981
outside
1 - 119
Population Genetic Test Statistics
Pi
110.088009
Theta
81.434742
Tajima's D
0.786618
CLR
0.168141
CSRT
0.5977201139943
Interpretation
Uncertain
