Gene
KWMTBOMO05190
Pre Gene Modal
BGIBMGA002445
Annotation
PREDICTED:_tRNA-specific_adenosine_deaminase_1-like_[Plutella_xylostella]
Full name
tRNA-specific adenosine deaminase 1
Alternative Name
dADAT1
tRNA-specific adenosine-37 deaminase
tRNA-specific adenosine-37 deaminase
Location in the cell
Nuclear Reliability : 3.832
Sequence
CDS
ATGCCAGATAACCTATCAAGTGTATGTGTCGATAACATAGTAGAGAAATGTTTGAAAACATACGAACAACTACCCAAAAAAGGAAAACCTGCCGATGACGAATGGACGGTGCTATCTTGCATTGTAAAATATGAAACTGAACATGATACGATCGAAGTATTATCCTTAGGAACTGGGTCTAAATGTATTGGCGCTACAAAAATGTCGCCACTCGGAGACTTGCTCAATGACAGTCACGCTGAAGTGTTCGCTCGACGCGGGTTTATACATTATCTATTCCAAAATATCGAAAAGGCTACCAACAATCTTGATTCTATATTCATTAAAACGGATAGCAAGCTAAAATTAAAAGACAGCATAGAATTTATATTTTATTCTTCACAATTACCTTGTGGTGACGCATCTATTATACCTAAGAATGGTGAAGAGCAAGAGGACCACTTTGGTGATCTAATCAAGGTTAAAAGAAAGGCAGATGAGAGCAACTGTGTGCGTGACACTAAAAGACTAAAATTAAGTGATATTCATCGAACCGGTGCAAAATGCCTTTCAAATGCAGAACAGGACCTTAAAATTCCTGGAAAAGATTATCATGTTTTAGGTCAGGTTAGAACTAAGCCTGGAAGAGGTGATAGAACACTATCAGTGTCTTGTAGTGATAAAATAGCTAGATATAAAATCAGAAGGAAAGTATAG
Protein
MPDNLSSVCVDNIVEKCLKTYEQLPKKGKPADDEWTVLSCIVKYETEHDTIEVLSLGTGSKCIGATKMSPLGDLLNDSHAEVFARRGFIHYLFQNIEKATNNLDSIFIKTDSKLKLKDSIEFIFYSSQLPCGDASIIPKNGEEQEDHFGDLIKVKRKADESNCVRDTKRLKLSDIHRTGAKCLSNAEQDLKIPGKDYHVLGQVRTKPGRGDRTLSVSCSDKIARYKIRRKV
Summary
Description
Specifically deaminates adenosine-37 to inosine in tRNA-Ala.
Catalytic Activity
adenosine(37) in tRNA(Ala) + H(+) + H2O = inosine(37) in tRNA(Ala) + NH4(+)
Cofactor
1D-myo-inositol hexakisphosphate
Similarity
Belongs to the ADAT1 family.
Keywords
Complete proteome
Hydrolase
Metal-binding
Reference proteome
tRNA processing
Zinc
Feature
chain tRNA-specific adenosine deaminase 1
Uniprot
H9IYW0
A0A3S2TNV0
A0A2H1VND2
A0A194RNF8
A0A2H1VL42
A0A0L7KTY9
+ More
A0A194PMS8 A0A0P6F8I2 A0A0P5H778 A0A0P4YKN3 A0A0P4Y4S7 A0A0P4XKQ2 A0A162PL27 A0A0P5PZK5 A0A0P6JJ14 E9GMF4 E0VK95 A0A3Q4GNB2 A0A146LV80 A0A139WKA0 V5GVN5 A0A1A8LYB7 U4UNI0 N6UDH2 A0A3P9CEE3 A0A3Q3D0V6 A0A2H8TWE4 A0A3Q3JCJ9 J3JVX3 A0A1A8F051 A0A0P5KPH6 A0A1B6K858 A0A3P8NUY1 A0A2S2NEH6 A0A1A8PKI9 G3NBY6 A0A1B6LNE2 A0A3B4TFS5 A0A1A8V942 A0A1A7ZQI5 A0A1A8IW17 J9LSD3 A0A3P8X4I3 A0A1A7XDU9 A0A3Q1CHG3 A0A3B4F681 A0A1B6H2C4 A0A3P8SCZ3 A0A3B4WLI3 A0A3Q3LMF6 A0A3B4B9H1 H2MJY2 A0A3Q3LMH3 K7J168 A0A3P9IUJ9 A0A0T6AV05 A0A3P9KVI8 A0A0C9R2C9 A0A3Q3GTJ7 A0A2R5LK74 A0A3Q1HM40 A0A131XZ26 A0A232FJ58 A0A224Z8T9 A0A1I8PCS3 A0A3P9N4V8 A0A2L2YHZ0 A0A3P9N4U8 F4X2X4 A0A1I8N125 A0A0P5QLZ8 A0A0P6D0B2 A0A0P5Z7D8 A0A3P9A0H3 A0A151X5H3 E9IK04 E2AUE8 B4P3Q0 A0A154PDH2 A0A182MM46 E2B4R0 B4HX96 B4Q560 A0A3L8DQF2 A0A293M997 A0A147BPZ1 A0A0L8GUP4 A0A195FSB2 N0BSY1 M9PDF5 Q9V3R6 A0A0L0CA60 A0A088ADX9 A0A0L7RG36 A0A3Q2DTE6 A0A182WH76 A0A3B1KET4 A0A1A9WCD8 A0A0P4W7T8
A0A194PMS8 A0A0P6F8I2 A0A0P5H778 A0A0P4YKN3 A0A0P4Y4S7 A0A0P4XKQ2 A0A162PL27 A0A0P5PZK5 A0A0P6JJ14 E9GMF4 E0VK95 A0A3Q4GNB2 A0A146LV80 A0A139WKA0 V5GVN5 A0A1A8LYB7 U4UNI0 N6UDH2 A0A3P9CEE3 A0A3Q3D0V6 A0A2H8TWE4 A0A3Q3JCJ9 J3JVX3 A0A1A8F051 A0A0P5KPH6 A0A1B6K858 A0A3P8NUY1 A0A2S2NEH6 A0A1A8PKI9 G3NBY6 A0A1B6LNE2 A0A3B4TFS5 A0A1A8V942 A0A1A7ZQI5 A0A1A8IW17 J9LSD3 A0A3P8X4I3 A0A1A7XDU9 A0A3Q1CHG3 A0A3B4F681 A0A1B6H2C4 A0A3P8SCZ3 A0A3B4WLI3 A0A3Q3LMF6 A0A3B4B9H1 H2MJY2 A0A3Q3LMH3 K7J168 A0A3P9IUJ9 A0A0T6AV05 A0A3P9KVI8 A0A0C9R2C9 A0A3Q3GTJ7 A0A2R5LK74 A0A3Q1HM40 A0A131XZ26 A0A232FJ58 A0A224Z8T9 A0A1I8PCS3 A0A3P9N4V8 A0A2L2YHZ0 A0A3P9N4U8 F4X2X4 A0A1I8N125 A0A0P5QLZ8 A0A0P6D0B2 A0A0P5Z7D8 A0A3P9A0H3 A0A151X5H3 E9IK04 E2AUE8 B4P3Q0 A0A154PDH2 A0A182MM46 E2B4R0 B4HX96 B4Q560 A0A3L8DQF2 A0A293M997 A0A147BPZ1 A0A0L8GUP4 A0A195FSB2 N0BSY1 M9PDF5 Q9V3R6 A0A0L0CA60 A0A088ADX9 A0A0L7RG36 A0A3Q2DTE6 A0A182WH76 A0A3B1KET4 A0A1A9WCD8 A0A0P4W7T8
EC Number
3.5.4.34
Pubmed
19121390
26354079
26227816
21292972
20566863
25186727
+ More
26823975 18362917 19820115 23537049 22516182 24487278 25463417 17554307 20075255 28648823 28797301 26561354 21719571 25315136 25069045 21282665 20798317 17994087 17550304 22936249 30249741 29652888 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10629039 26108605 25329095
26823975 18362917 19820115 23537049 22516182 24487278 25463417 17554307 20075255 28648823 28797301 26561354 21719571 25315136 25069045 21282665 20798317 17994087 17550304 22936249 30249741 29652888 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10629039 26108605 25329095
EMBL
BABH01010679
RSAL01000038
RVE51145.1
ODYU01003479
SOQ42330.1
KQ459984
+ More
KPJ18949.1 ODYU01003150 SOQ41538.1 JTDY01005703 KOB66722.1 KQ459597 KPI94736.1 GDIQ01053975 JAN40762.1 GDIQ01230254 JAK21471.1 GDIP01226029 JAI97372.1 GDIP01233731 JAI89670.1 GDIP01241240 JAI82161.1 LRGB01000494 KZS18938.1 GDIQ01121896 JAL29830.1 GDIQ01006326 JAN88411.1 GL732552 EFX79359.1 DS235239 EEB13801.1 GDHC01008032 JAQ10597.1 KQ971328 KYB28362.1 GALX01002709 JAB65757.1 HAEF01010115 SBR49780.1 KB632292 ERL91575.1 APGK01026978 KB740648 ENN79750.1 GFXV01006762 MBW18567.1 BT127391 AEE62353.1 HAEB01005960 SBQ52487.1 GDIQ01186566 JAK65159.1 GECU01000112 JAT07595.1 GGMR01002928 MBY15547.1 HAEH01007302 SBR81527.1 GEBQ01014725 JAT25252.1 HAEJ01015884 SBS56341.1 HADY01006071 SBP44556.1 HAED01015087 SBR01532.1 ABLF02040844 HADW01014768 SBP16168.1 GECZ01000999 JAS68770.1 LJIG01022802 KRT78638.1 GBYB01002149 GBYB01002150 JAG71916.1 JAG71917.1 GGLE01005753 MBY09879.1 GEFM01003537 JAP72259.1 NNAY01000154 OXU30498.1 GFPF01012885 MAA24031.1 IAAA01029427 LAA07761.1 GL888600 EGI59195.1 GDIQ01133036 JAL18690.1 GDIQ01087094 JAN07643.1 GDIP01047857 JAM55858.1 KQ982498 KYQ55651.1 GL763921 EFZ19089.1 GL442815 EFN63005.1 CM000157 EDW88351.1 KQ434878 KZC09857.1 AXCM01017536 GL445584 EFN89341.1 CH480818 EDW51676.1 CM000361 CM002910 EDX04966.1 KMY90159.1 QOIP01000005 RLU22670.1 GFWV01012020 MAA36749.1 GEGO01002576 JAR92828.1 KQ420350 KOF80554.1 KQ981280 KYN43353.1 BT150087 AGK63853.1 AE014134 AGB92985.1 AF192530 JRES01000685 KNC29142.1 KQ414596 KOC69912.1 GDRN01084205 JAI61534.1
KPJ18949.1 ODYU01003150 SOQ41538.1 JTDY01005703 KOB66722.1 KQ459597 KPI94736.1 GDIQ01053975 JAN40762.1 GDIQ01230254 JAK21471.1 GDIP01226029 JAI97372.1 GDIP01233731 JAI89670.1 GDIP01241240 JAI82161.1 LRGB01000494 KZS18938.1 GDIQ01121896 JAL29830.1 GDIQ01006326 JAN88411.1 GL732552 EFX79359.1 DS235239 EEB13801.1 GDHC01008032 JAQ10597.1 KQ971328 KYB28362.1 GALX01002709 JAB65757.1 HAEF01010115 SBR49780.1 KB632292 ERL91575.1 APGK01026978 KB740648 ENN79750.1 GFXV01006762 MBW18567.1 BT127391 AEE62353.1 HAEB01005960 SBQ52487.1 GDIQ01186566 JAK65159.1 GECU01000112 JAT07595.1 GGMR01002928 MBY15547.1 HAEH01007302 SBR81527.1 GEBQ01014725 JAT25252.1 HAEJ01015884 SBS56341.1 HADY01006071 SBP44556.1 HAED01015087 SBR01532.1 ABLF02040844 HADW01014768 SBP16168.1 GECZ01000999 JAS68770.1 LJIG01022802 KRT78638.1 GBYB01002149 GBYB01002150 JAG71916.1 JAG71917.1 GGLE01005753 MBY09879.1 GEFM01003537 JAP72259.1 NNAY01000154 OXU30498.1 GFPF01012885 MAA24031.1 IAAA01029427 LAA07761.1 GL888600 EGI59195.1 GDIQ01133036 JAL18690.1 GDIQ01087094 JAN07643.1 GDIP01047857 JAM55858.1 KQ982498 KYQ55651.1 GL763921 EFZ19089.1 GL442815 EFN63005.1 CM000157 EDW88351.1 KQ434878 KZC09857.1 AXCM01017536 GL445584 EFN89341.1 CH480818 EDW51676.1 CM000361 CM002910 EDX04966.1 KMY90159.1 QOIP01000005 RLU22670.1 GFWV01012020 MAA36749.1 GEGO01002576 JAR92828.1 KQ420350 KOF80554.1 KQ981280 KYN43353.1 BT150087 AGK63853.1 AE014134 AGB92985.1 AF192530 JRES01000685 KNC29142.1 KQ414596 KOC69912.1 GDRN01084205 JAI61534.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000037510
UP000053268
UP000076858
+ More
UP000000305 UP000009046 UP000261580 UP000007266 UP000030742 UP000019118 UP000265160 UP000264840 UP000261600 UP000265100 UP000007635 UP000261420 UP000007819 UP000265120 UP000257160 UP000261460 UP000265080 UP000261360 UP000261660 UP000261520 UP000001038 UP000002358 UP000265200 UP000265180 UP000265040 UP000215335 UP000095300 UP000242638 UP000007755 UP000095301 UP000265140 UP000075809 UP000000311 UP000002282 UP000076502 UP000075883 UP000008237 UP000001292 UP000000304 UP000279307 UP000053454 UP000078541 UP000000803 UP000037069 UP000005203 UP000053825 UP000265020 UP000075920 UP000018467 UP000091820
UP000000305 UP000009046 UP000261580 UP000007266 UP000030742 UP000019118 UP000265160 UP000264840 UP000261600 UP000265100 UP000007635 UP000261420 UP000007819 UP000265120 UP000257160 UP000261460 UP000265080 UP000261360 UP000261660 UP000261520 UP000001038 UP000002358 UP000265200 UP000265180 UP000265040 UP000215335 UP000095300 UP000242638 UP000007755 UP000095301 UP000265140 UP000075809 UP000000311 UP000002282 UP000076502 UP000075883 UP000008237 UP000001292 UP000000304 UP000279307 UP000053454 UP000078541 UP000000803 UP000037069 UP000005203 UP000053825 UP000265020 UP000075920 UP000018467 UP000091820
Interpro
SUPFAM
SSF51430
SSF51430
Gene 3D
ProteinModelPortal
H9IYW0
A0A3S2TNV0
A0A2H1VND2
A0A194RNF8
A0A2H1VL42
A0A0L7KTY9
+ More
A0A194PMS8 A0A0P6F8I2 A0A0P5H778 A0A0P4YKN3 A0A0P4Y4S7 A0A0P4XKQ2 A0A162PL27 A0A0P5PZK5 A0A0P6JJ14 E9GMF4 E0VK95 A0A3Q4GNB2 A0A146LV80 A0A139WKA0 V5GVN5 A0A1A8LYB7 U4UNI0 N6UDH2 A0A3P9CEE3 A0A3Q3D0V6 A0A2H8TWE4 A0A3Q3JCJ9 J3JVX3 A0A1A8F051 A0A0P5KPH6 A0A1B6K858 A0A3P8NUY1 A0A2S2NEH6 A0A1A8PKI9 G3NBY6 A0A1B6LNE2 A0A3B4TFS5 A0A1A8V942 A0A1A7ZQI5 A0A1A8IW17 J9LSD3 A0A3P8X4I3 A0A1A7XDU9 A0A3Q1CHG3 A0A3B4F681 A0A1B6H2C4 A0A3P8SCZ3 A0A3B4WLI3 A0A3Q3LMF6 A0A3B4B9H1 H2MJY2 A0A3Q3LMH3 K7J168 A0A3P9IUJ9 A0A0T6AV05 A0A3P9KVI8 A0A0C9R2C9 A0A3Q3GTJ7 A0A2R5LK74 A0A3Q1HM40 A0A131XZ26 A0A232FJ58 A0A224Z8T9 A0A1I8PCS3 A0A3P9N4V8 A0A2L2YHZ0 A0A3P9N4U8 F4X2X4 A0A1I8N125 A0A0P5QLZ8 A0A0P6D0B2 A0A0P5Z7D8 A0A3P9A0H3 A0A151X5H3 E9IK04 E2AUE8 B4P3Q0 A0A154PDH2 A0A182MM46 E2B4R0 B4HX96 B4Q560 A0A3L8DQF2 A0A293M997 A0A147BPZ1 A0A0L8GUP4 A0A195FSB2 N0BSY1 M9PDF5 Q9V3R6 A0A0L0CA60 A0A088ADX9 A0A0L7RG36 A0A3Q2DTE6 A0A182WH76 A0A3B1KET4 A0A1A9WCD8 A0A0P4W7T8
A0A194PMS8 A0A0P6F8I2 A0A0P5H778 A0A0P4YKN3 A0A0P4Y4S7 A0A0P4XKQ2 A0A162PL27 A0A0P5PZK5 A0A0P6JJ14 E9GMF4 E0VK95 A0A3Q4GNB2 A0A146LV80 A0A139WKA0 V5GVN5 A0A1A8LYB7 U4UNI0 N6UDH2 A0A3P9CEE3 A0A3Q3D0V6 A0A2H8TWE4 A0A3Q3JCJ9 J3JVX3 A0A1A8F051 A0A0P5KPH6 A0A1B6K858 A0A3P8NUY1 A0A2S2NEH6 A0A1A8PKI9 G3NBY6 A0A1B6LNE2 A0A3B4TFS5 A0A1A8V942 A0A1A7ZQI5 A0A1A8IW17 J9LSD3 A0A3P8X4I3 A0A1A7XDU9 A0A3Q1CHG3 A0A3B4F681 A0A1B6H2C4 A0A3P8SCZ3 A0A3B4WLI3 A0A3Q3LMF6 A0A3B4B9H1 H2MJY2 A0A3Q3LMH3 K7J168 A0A3P9IUJ9 A0A0T6AV05 A0A3P9KVI8 A0A0C9R2C9 A0A3Q3GTJ7 A0A2R5LK74 A0A3Q1HM40 A0A131XZ26 A0A232FJ58 A0A224Z8T9 A0A1I8PCS3 A0A3P9N4V8 A0A2L2YHZ0 A0A3P9N4U8 F4X2X4 A0A1I8N125 A0A0P5QLZ8 A0A0P6D0B2 A0A0P5Z7D8 A0A3P9A0H3 A0A151X5H3 E9IK04 E2AUE8 B4P3Q0 A0A154PDH2 A0A182MM46 E2B4R0 B4HX96 B4Q560 A0A3L8DQF2 A0A293M997 A0A147BPZ1 A0A0L8GUP4 A0A195FSB2 N0BSY1 M9PDF5 Q9V3R6 A0A0L0CA60 A0A088ADX9 A0A0L7RG36 A0A3Q2DTE6 A0A182WH76 A0A3B1KET4 A0A1A9WCD8 A0A0P4W7T8
PDB
6D06
E-value=9.675e-16,
Score=201
Ontologies
GO
PANTHER
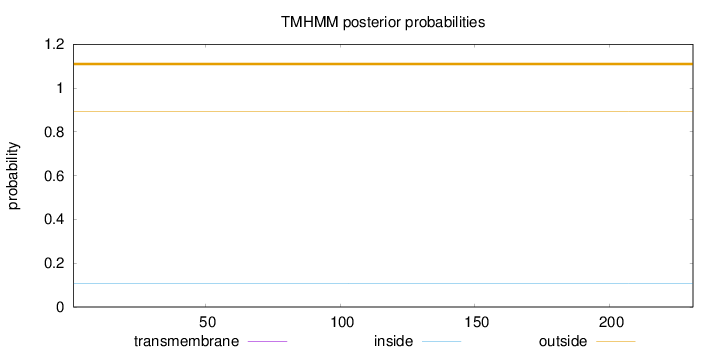
Topology
Length:
231
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00013
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.10580
outside
1 - 231
Population Genetic Test Statistics
Pi
185.993151
Theta
148.542177
Tajima's D
0.805195
CLR
0
CSRT
0.607469626518674
Interpretation
Uncertain
