Gene
KWMTBOMO05184
Pre Gene Modal
BGIBMGA002451
Annotation
PREDICTED:_disks_large_homolog_2-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.015
Sequence
CDS
ATGACCACGTGGATGGCGCTGTTTGGGCTAGTGTGGCTGCTGCGAGTGCGCGGCTCCCTCTGTGGTTGTGCCGCGTGCAAGCGCGTCAGACACGAGACCACCGAGATCCCGAACGTTAGCGCAGCCTCTCAGAGTTCACCAACAGAAGAACGGGCGTCTTTTGACGTCATCGCGCTACAGCACTACACTCGTGACGTCACCGTACAAACTGACTTCGAGGACGAAGAGATCGAGGACCTCGATAAGATCGAACTCCAAATATCTGAGGAATCTAACGTGGGTAACTATGAGTGCGGCCGAGAGCAGCCTGCACAAAGTCCCGGAAACGCCCGACGTTCTGCAGGAAGCTACCAGTACACTTCAGAAGCGGACGAGTCTGACTGGGAAACTTGTGACGTAACGCTGGAGCGGGGTGCGAGTGGTCTTGGGCTTAGCATAGCGGGCGGCGAGACAGACGGCGACGTCACAATCACCAGACTAGCAGCCGGAGGGGCAGCCAAGAAAGATGGCAGACTACAAATAGGCGATGTTTTATTGCAAGTTAATGACATCTCCGTGGAGGGTGCGTCGCATTCTGTCGCAGTGGATGCATTGCAAAAAGCGGGCAACGTTGTCAGATTACGAGTTTGCCGTGCCCGAAGACCAAGAGTGGTGTCGTTGTGGCGTGGCGTTCGCGGCCTCGGACTCGGTATAGCGGGAGGGTCTGATGACGCGGCTGGAGGAGGAGGCGTTTTTATATCACATATAGCCGTCGGTGGTGCAGCACATCACGACGGCAGACTGAGATTAGGCGATAAAATACTTGCAGTCAGGGATGAAGATGGAATCGAGACATCGCTTGTTGGTGCGACTCACGCACAAGCAGTGTCTGCACTTCGGAATACAGGGGAGCAAGTTACCTTAGTAGTATTACCAGCCGGTTCGGTGCCGCCCGTTGCCAAAACCGCACCGCTTTACTCTACGAGAACACAAGCTACTTCGTGCTCAACACTACACGAACTTCTTGAGGAGGAGCCAAGTGAAATACCGAGATGCGTTCGGATGGTTCGCTTGGTCCGTTCTGGATCGCGGCTAGGTATGGACATCGTCGGCGGTCTTGGCGGTGACTCTGAAGGGATTGAAGGCGAGGAGGATACTTGTGGGGTATTTGTTTCGGGAGTATCCGTGGCGGGAGCAGCTTACGGTATGCTGCATAGAGGGGACAGGATACTCAGCGTTGATGGACGTGATTTAACAAGAGCCACTCATGAACAAGCTGCCGCTGCGTTGAAGTATTCTGGGAGCGCAGTGACGATAGCTGCACAATACCAACCGGAGCAGTATGAAAGACTAAGGGCGCGGATACGAGCAATCAATGCAGCTGCGATGTCGCCTCACGCCGTACATGCCACCCATGTTCCTGTACATCCGGACTTACACACCATGTACCCAAGGTAG
Protein
MTTWMALFGLVWLLRVRGSLCGCAACKRVRHETTEIPNVSAASQSSPTEERASFDVIALQHYTRDVTVQTDFEDEEIEDLDKIELQISEESNVGNYECGREQPAQSPGNARRSAGSYQYTSEADESDWETCDVTLERGASGLGLSIAGGETDGDVTITRLAAGGAAKKDGRLQIGDVLLQVNDISVEGASHSVAVDALQKAGNVVRLRVCRARRPRVVSLWRGVRGLGLGIAGGSDDAAGGGGVFISHIAVGGAAHHDGRLRLGDKILAVRDEDGIETSLVGATHAQAVSALRNTGEQVTLVVLPAGSVPPVAKTAPLYSTRTQATSCSTLHELLEEEPSEIPRCVRMVRLVRSGSRLGMDIVGGLGGDSEGIEGEEDTCGVFVSGVSVAGAAYGMLHRGDRILSVDGRDLTRATHEQAAAALKYSGSAVTIAAQYQPEQYERLRARIRAINAAAMSPHAVHATHVPVHPDLHTMYPR
Summary
Similarity
Belongs to the MAGUK family.
Uniprot
H9IYW6
A0A194PDT1
A0A194RM91
A0A212EKQ4
V9KR91
V9KAM2
+ More
V9KFQ4 A0A1A7YTE1 A0A1A7WHR7 A0A1A7W9T0 A0A1A8LLT6 A0A1A8R065 A0A1A8C351 A0A1A8V4N0 A0A1A8ICM9 A0A1A8LUV8 A0A1A8BT75 A0A1A8R3Q6 A0A1A8AGJ7 A0A1A8QFJ0 A0A1A8IDS8 A0A1A8R0S3 A0A3B3DCI5 A0A3P9LX49 H2M6D5 A0A3B3DBX9 A0A1A8FNZ4 A0A1A8GQ52 A0A3B5RCU9 A0A3P9KC95 A0A1A8FKD0 A0A3B3HSS3 A0A3B3DDG3 A0A3B3HNI5 A0A3B3DDS7 A0A3P9LWS6 A0A3B5Q4A1 A0A3B3DCG3 M3ZW70 A0A3B3I950 A0A3B3HAW1 A0A3P9LX19 A0A3B3BW68 A0A3B3HW66 A0A3P9LXI5 A0A3P9LX12 A0A3B3BVL1 A0A3P9LWQ7 A0A2I4BZP4 A0A3Q3W1C2 A0A3B3DC90 A0A3P9LWH4 A0A3B3HRQ2 A0A3Q3VUU4 A0A3Q3VS73 A0A3Q3VUP8 A0A2I4BZP6 A0A3B3DC69 A0A2I4BZQ1 A0A3Q4ALW2 A0A2I4BZT5 A0A2I4BZQ2 A0A3P9H7I0 A0A3P9H7W9 A0A3P9IVR5
V9KFQ4 A0A1A7YTE1 A0A1A7WHR7 A0A1A7W9T0 A0A1A8LLT6 A0A1A8R065 A0A1A8C351 A0A1A8V4N0 A0A1A8ICM9 A0A1A8LUV8 A0A1A8BT75 A0A1A8R3Q6 A0A1A8AGJ7 A0A1A8QFJ0 A0A1A8IDS8 A0A1A8R0S3 A0A3B3DCI5 A0A3P9LX49 H2M6D5 A0A3B3DBX9 A0A1A8FNZ4 A0A1A8GQ52 A0A3B5RCU9 A0A3P9KC95 A0A1A8FKD0 A0A3B3HSS3 A0A3B3DDG3 A0A3B3HNI5 A0A3B3DDS7 A0A3P9LWS6 A0A3B5Q4A1 A0A3B3DCG3 M3ZW70 A0A3B3I950 A0A3B3HAW1 A0A3P9LX19 A0A3B3BW68 A0A3B3HW66 A0A3P9LXI5 A0A3P9LX12 A0A3B3BVL1 A0A3P9LWQ7 A0A2I4BZP4 A0A3Q3W1C2 A0A3B3DC90 A0A3P9LWH4 A0A3B3HRQ2 A0A3Q3VUU4 A0A3Q3VS73 A0A3Q3VUP8 A0A2I4BZP6 A0A3B3DC69 A0A2I4BZQ1 A0A3Q4ALW2 A0A2I4BZT5 A0A2I4BZQ2 A0A3P9H7I0 A0A3P9H7W9 A0A3P9IVR5
EMBL
BABH01010635
KQ459606
KPI91417.1
KQ459984
KPJ18943.1
AGBW02014221
+ More
OWR42064.1 JW868370 AFP00888.1 JW862178 AFO94695.1 JW864311 AFO96828.1 HADX01011229 SBP33461.1 HADW01003885 SBP05285.1 HADW01001203 SBP02603.1 HAEF01007477 SBR44859.1 HAEH01014174 SBR99316.1 HADZ01010043 HAEA01011237 SBP73984.1 HADY01016008 HAEJ01014853 SBS55310.1 HAED01008427 HAEE01012156 SBQ94639.1 HAEF01009235 SBR48106.1 HADZ01006926 SBP70867.1 HAEG01015609 SBS00601.1 HADY01015767 SBP54252.1 HAEI01005185 SBR92152.1 HAED01009006 SBQ95218.1 HAEH01014072 SBR99053.1 HAEB01014046 SBQ60573.1 HAEC01005898 SBQ73975.1 HAEB01012915 SBQ59442.1
OWR42064.1 JW868370 AFP00888.1 JW862178 AFO94695.1 JW864311 AFO96828.1 HADX01011229 SBP33461.1 HADW01003885 SBP05285.1 HADW01001203 SBP02603.1 HAEF01007477 SBR44859.1 HAEH01014174 SBR99316.1 HADZ01010043 HAEA01011237 SBP73984.1 HADY01016008 HAEJ01014853 SBS55310.1 HAED01008427 HAEE01012156 SBQ94639.1 HAEF01009235 SBR48106.1 HADZ01006926 SBP70867.1 HAEG01015609 SBS00601.1 HADY01015767 SBP54252.1 HAEI01005185 SBR92152.1 HAED01009006 SBQ95218.1 HAEH01014072 SBR99053.1 HAEB01014046 SBQ60573.1 HAEC01005898 SBQ73975.1 HAEB01012915 SBQ59442.1
Proteomes
PRIDE
Pfam
Interpro
IPR001478
PDZ
+ More
IPR036034 PDZ_sf
IPR019590 DLG1_PEST_dom
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR019583 PDZ_assoc
IPR008144 Guanylate_kin-like_dom
IPR008145 GK/Ca_channel_bsu
IPR016313 DLG1-like
IPR020590 Guanylate_kinase_CS
IPR027417 P-loop_NTPase
IPR015143 L27_1
IPR004172 L27_dom
IPR036892 L27_dom_sf
IPR036034 PDZ_sf
IPR019590 DLG1_PEST_dom
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR019583 PDZ_assoc
IPR008144 Guanylate_kin-like_dom
IPR008145 GK/Ca_channel_bsu
IPR016313 DLG1-like
IPR020590 Guanylate_kinase_CS
IPR027417 P-loop_NTPase
IPR015143 L27_1
IPR004172 L27_dom
IPR036892 L27_dom_sf
ProteinModelPortal
H9IYW6
A0A194PDT1
A0A194RM91
A0A212EKQ4
V9KR91
V9KAM2
+ More
V9KFQ4 A0A1A7YTE1 A0A1A7WHR7 A0A1A7W9T0 A0A1A8LLT6 A0A1A8R065 A0A1A8C351 A0A1A8V4N0 A0A1A8ICM9 A0A1A8LUV8 A0A1A8BT75 A0A1A8R3Q6 A0A1A8AGJ7 A0A1A8QFJ0 A0A1A8IDS8 A0A1A8R0S3 A0A3B3DCI5 A0A3P9LX49 H2M6D5 A0A3B3DBX9 A0A1A8FNZ4 A0A1A8GQ52 A0A3B5RCU9 A0A3P9KC95 A0A1A8FKD0 A0A3B3HSS3 A0A3B3DDG3 A0A3B3HNI5 A0A3B3DDS7 A0A3P9LWS6 A0A3B5Q4A1 A0A3B3DCG3 M3ZW70 A0A3B3I950 A0A3B3HAW1 A0A3P9LX19 A0A3B3BW68 A0A3B3HW66 A0A3P9LXI5 A0A3P9LX12 A0A3B3BVL1 A0A3P9LWQ7 A0A2I4BZP4 A0A3Q3W1C2 A0A3B3DC90 A0A3P9LWH4 A0A3B3HRQ2 A0A3Q3VUU4 A0A3Q3VS73 A0A3Q3VUP8 A0A2I4BZP6 A0A3B3DC69 A0A2I4BZQ1 A0A3Q4ALW2 A0A2I4BZT5 A0A2I4BZQ2 A0A3P9H7I0 A0A3P9H7W9 A0A3P9IVR5
V9KFQ4 A0A1A7YTE1 A0A1A7WHR7 A0A1A7W9T0 A0A1A8LLT6 A0A1A8R065 A0A1A8C351 A0A1A8V4N0 A0A1A8ICM9 A0A1A8LUV8 A0A1A8BT75 A0A1A8R3Q6 A0A1A8AGJ7 A0A1A8QFJ0 A0A1A8IDS8 A0A1A8R0S3 A0A3B3DCI5 A0A3P9LX49 H2M6D5 A0A3B3DBX9 A0A1A8FNZ4 A0A1A8GQ52 A0A3B5RCU9 A0A3P9KC95 A0A1A8FKD0 A0A3B3HSS3 A0A3B3DDG3 A0A3B3HNI5 A0A3B3DDS7 A0A3P9LWS6 A0A3B5Q4A1 A0A3B3DCG3 M3ZW70 A0A3B3I950 A0A3B3HAW1 A0A3P9LX19 A0A3B3BW68 A0A3B3HW66 A0A3P9LXI5 A0A3P9LX12 A0A3B3BVL1 A0A3P9LWQ7 A0A2I4BZP4 A0A3Q3W1C2 A0A3B3DC90 A0A3P9LWH4 A0A3B3HRQ2 A0A3Q3VUU4 A0A3Q3VS73 A0A3Q3VUP8 A0A2I4BZP6 A0A3B3DC69 A0A2I4BZQ1 A0A3Q4ALW2 A0A2I4BZT5 A0A2I4BZQ2 A0A3P9H7I0 A0A3P9H7W9 A0A3P9IVR5
PDB
2XKX
E-value=1.81764e-25,
Score=288
Ontologies
GO
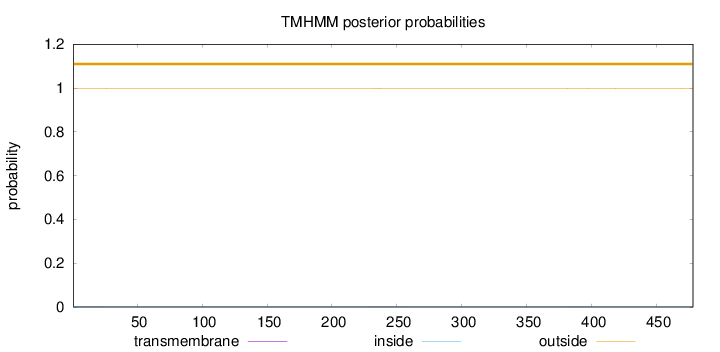
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.845065
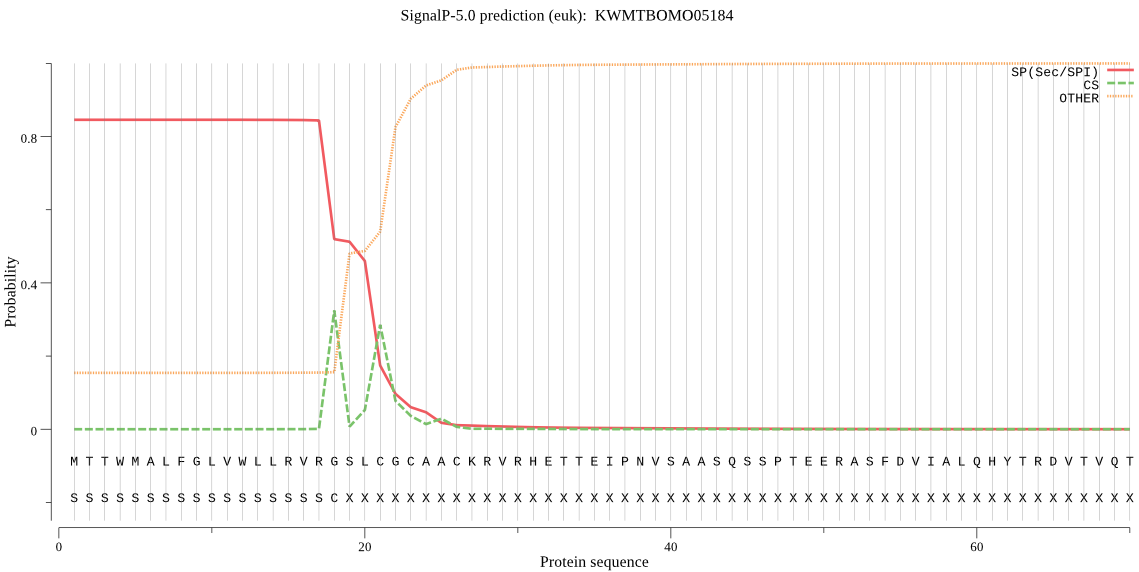
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0379000000000001
Exp number, first 60 AAs:
0.01392
Total prob of N-in:
0.00137
outside
1 - 478
Population Genetic Test Statistics
Pi
241.024432
Theta
195.046338
Tajima's D
0.579638
CLR
0.28423
CSRT
0.533673316334183
Interpretation
Uncertain
