Gene
KWMTBOMO05183
Pre Gene Modal
BGIBMGA002450
Annotation
PREDICTED:_myrosinase_1-like_[Plutella_xylostella]
Location in the cell
Peroxisomal Reliability : 0.965
Sequence
CDS
ATGCGGAGCTCGCGTAGTGTCGCCTTAAGCGGTGTAGTGTTAGTAACACTAAGCTTGCTGTCGTGTTCACATGAAGCAGTGGTGTCTGATATCTATTACGGGGTGAGCAATTACTCCTTTCCTGAGGATTTTATATTCGGTGTCTCGACGGCCGCATTTCAAATAGAAGGAGCGTGGAATGAGGGAGGAAAAGGAGAGAGTATTTGGGACACGTACGTTCACGAGCATCCGGAATATACCCCCGATAAGTCGAATGGAGATATTGCAGCTGATTCATATCACAAATTTAAAGAAGACGTACAGTTGATTAAGGCACTGGGAGTCAAATATTATCGTCTGTCAATATCGTGGCCGCGCTTGCTACCAAATGGAACAAGCAATAACATAAGTACTGATGGAGCGCGATACTATCGAAATCTTTTCGAAGAGCTTTTGAAAGCGAATATAACTCCTATTGTTACGTTATTCCATTGGGATATGCCGACGCCCTTTATGGACCTAGGAGGATGGAACAACCCCGTGATAGTGGATTACTTCGTAGATTACGCAAGGGTCGCGTTCAATTTGTACGGTGACATCATTAAAACGTGGACCACAATTAACGAGCCACATCAGCATTGTTACCAAGGTTACGGAGCCGACTACTTCGCACCAGCCATAAAGTCACACGGTGTAGGAGAATATTTATGTGCACACTATTTAATCTTAAGTCACGCAAAAGCATATCATTTATACGAAAAAGAATTCAAGCCGCATCAAAAAGGTAAAGTTGGAATTACTTTAGATGCATTTATGGGAATACCAAAGGATCCTCGGAACTTTCAAGATGTACTAGCTGTTCGAAGTTACCTGCAAATTCATTTTGATCTTTACGCTCATCCAATCTTTTCTGCCGAGGGAGATTATCCTACATTCGTGCGACAAAGAATTAACAATATGAGCTTGGCTCAGGGATTTGCAAGATCACGTTTACCGTATTTTACTGCAGAAGAGGTCAAAATAATCCGCGGCAGCTCAGATTTCTTCGGACTTAACCATTATACCACGTACTTGATGTCACCATCTGAAATGGAATTTTCCTGGTCGATACCTTCAATGGATCACGATGCTCAAGTTAGAATGGAACAAGACCCACACTGGGAAAAACCCGGTACAGACTGGCTTTCGGTTTACCCAGAAGGGTTTCGAAAGTTATTGAACTACGTAAGCGATAATTACGGAAGAGAGATTCCTATAATTGTCACAGAAAATGGCGTCGGTGACAAAGGGCAACTAAAGGACTACGATAGGATCTCATACTTCAACAAATACCTATATCAATTGCTTTTAGCCATACACGAGGACAACTGTAACATTATAGGATATTTTGCTTGGACCCTTATGGACGACTTTGAATGGTCAGATGGATACTTGTCGAAGTTTGGGCTATACAAAGTGGACTTTGACAGCCCTGAAAGAACCCGTACACCAAAACTCTCAGTATCGAGCTACCGCAACATAGTACAAACTAGAAGAATTAATTTTGACCGGTACAAGCCGCTTCCGAATAAACTCAATCATTTTTGA
Protein
MRSSRSVALSGVVLVTLSLLSCSHEAVVSDIYYGVSNYSFPEDFIFGVSTAAFQIEGAWNEGGKGESIWDTYVHEHPEYTPDKSNGDIAADSYHKFKEDVQLIKALGVKYYRLSISWPRLLPNGTSNNISTDGARYYRNLFEELLKANITPIVTLFHWDMPTPFMDLGGWNNPVIVDYFVDYARVAFNLYGDIIKTWTTINEPHQHCYQGYGADYFAPAIKSHGVGEYLCAHYLILSHAKAYHLYEKEFKPHQKGKVGITLDAFMGIPKDPRNFQDVLAVRSYLQIHFDLYAHPIFSAEGDYPTFVRQRINNMSLAQGFARSRLPYFTAEEVKIIRGSSDFFGLNHYTTYLMSPSEMEFSWSIPSMDHDAQVRMEQDPHWEKPGTDWLSVYPEGFRKLLNYVSDNYGREIPIIVTENGVGDKGQLKDYDRISYFNKYLYQLLLAIHEDNCNIIGYFAWTLMDDFEWSDGYLSKFGLYKVDFDSPERTRTPKLSVSSYRNIVQTRRINFDRYKPLPNKLNHF
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Uniprot
H9IYW5
G9F9I2
A0A212EKQ3
B2DBM6
A0A3S2TR57
A0A0K2YR16
+ More
A0A2A4IUI6 A0A194PDJ4 A0A2A4ISU7 A0A194RN51 H9IZH5 A0A2A4JTU1 A0A194PVV9 A0A1W1EGM5 A0A2H1VWC6 A0A3S2M0F9 A0A182J2M6 A0A194R738 B0WNN7 A0A219WGQ6 A0A182YP84 B4MM56 A0A182QNT7 A0A182HAK7 A0A1A9W4X6 A0A1Q3FY94 A0A1Y9IW73 Q16WF2 A0A1W4XAC4 A0A0A1XS57 A0A182QI13 A0A1S4G3W5 A0A2M4CLR8 A0A0A1XAG3 Q16ET6 A0A034VAL4 A0A1A9UP13 A0A0T6B9C6 A0A034VEY7 A0A1B0C2P0 A0A1W4UV58 A0A1A9XR01 A0A1A9ZLX0 A0A0K8UIW8 W5JPP3 Q5TQY8 A0A182LL18 A0A1Y9IRY8 A0A1Y9GKT0 A0A182U0B4 A0A0K8W4X1 A0A1Y9IVZ0 A0A212ERQ2 Q7PI98 T1PC49 A0A0M4F1P7 B3M4E4 B4KXZ5 A0A0K8W3Q7 A0A0M4F0Y9 A0A0J9RWT3 A0A1Q3FR87 A0A2M4CZ21 B4PKA3 A0A182TWE4 B4MM55 A0A182LWK9 A0A1Y9IW90 A0A1Y9GL87 B4HK62 Q9VV98 A0A1Y9J0J6 W8BL15 B3NDI0 A0A1L8DQN1 B4QMY1 A0A2M3ZET6 B4IXF6 J9Z300 B4LG53 A0A2M4BKN2 A0A2J7PTJ0 W5JN46 A0A1I8PM40 A0A1Y9IS75 A0A2J7PKH8 A0A3S2PE71 A0A084VCJ7 A0A182YP82 A0A067R142 W5JKR6 A0A067RC84 A0A182JLR1 A0A1B6D5W8 A0A1L8DR18 A0A1Y9IRG1 A0A1L8DQS5 G9F9I1 A0A1Y9J0G9 B4H7D1
A0A2A4IUI6 A0A194PDJ4 A0A2A4ISU7 A0A194RN51 H9IZH5 A0A2A4JTU1 A0A194PVV9 A0A1W1EGM5 A0A2H1VWC6 A0A3S2M0F9 A0A182J2M6 A0A194R738 B0WNN7 A0A219WGQ6 A0A182YP84 B4MM56 A0A182QNT7 A0A182HAK7 A0A1A9W4X6 A0A1Q3FY94 A0A1Y9IW73 Q16WF2 A0A1W4XAC4 A0A0A1XS57 A0A182QI13 A0A1S4G3W5 A0A2M4CLR8 A0A0A1XAG3 Q16ET6 A0A034VAL4 A0A1A9UP13 A0A0T6B9C6 A0A034VEY7 A0A1B0C2P0 A0A1W4UV58 A0A1A9XR01 A0A1A9ZLX0 A0A0K8UIW8 W5JPP3 Q5TQY8 A0A182LL18 A0A1Y9IRY8 A0A1Y9GKT0 A0A182U0B4 A0A0K8W4X1 A0A1Y9IVZ0 A0A212ERQ2 Q7PI98 T1PC49 A0A0M4F1P7 B3M4E4 B4KXZ5 A0A0K8W3Q7 A0A0M4F0Y9 A0A0J9RWT3 A0A1Q3FR87 A0A2M4CZ21 B4PKA3 A0A182TWE4 B4MM55 A0A182LWK9 A0A1Y9IW90 A0A1Y9GL87 B4HK62 Q9VV98 A0A1Y9J0J6 W8BL15 B3NDI0 A0A1L8DQN1 B4QMY1 A0A2M3ZET6 B4IXF6 J9Z300 B4LG53 A0A2M4BKN2 A0A2J7PTJ0 W5JN46 A0A1I8PM40 A0A1Y9IS75 A0A2J7PKH8 A0A3S2PE71 A0A084VCJ7 A0A182YP82 A0A067R142 W5JKR6 A0A067RC84 A0A182JLR1 A0A1B6D5W8 A0A1L8DR18 A0A1Y9IRG1 A0A1L8DQS5 G9F9I1 A0A1Y9J0G9 B4H7D1
Pubmed
19121390
22118469
18712529
26940001
26354079
28680679
+ More
25244985 17994087 26483478 17510324 25830018 25348373 20920257 23761445 12364791 14747013 17210077 20966253 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 24438588 24845553
25244985 17994087 26483478 17510324 25830018 25348373 20920257 23761445 12364791 14747013 17210077 20966253 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 24438588 24845553
EMBL
BABH01010632
BABH01010633
JN033727
AEW46880.1
AGBW02014221
OWR42063.1
+ More
AB264710 BAG30785.1 RSAL01000018 RVE52880.1 LN851681 CRL08768.1 NWSH01007170 PCG63058.1 KQ459606 KPI91416.1 PCG63057.1 KQ459984 KPJ18942.1 BABH01014157 NWSH01000598 PCG75431.1 KQ459589 KPI97521.1 LT635664 SGZ49394.1 ODYU01004836 SOQ45125.1 RSAL01000086 RVE48281.1 KQ460870 KPJ11681.1 DS232013 EDS31873.1 KT992464 AOY34571.1 CH963847 EDW73065.2 AXCN02000455 JXUM01031352 KQ560919 KXJ80348.1 GFDL01002519 JAV32526.1 CH477568 EAT38910.1 GBXI01000924 JAD13368.1 GGFL01002037 MBW66215.1 GBXI01006185 JAD08107.1 CH478582 EAT32750.1 GAKP01018616 JAC40336.1 LJIG01003002 KRT83938.1 GAKP01018617 JAC40335.1 JXJN01024604 JXJN01024605 JXJN01024606 GDHF01025685 JAI26629.1 ADMH02000892 ETN64734.1 AAAB01008960 EAL40075.2 APCN01002262 GDHF01006374 JAI45940.1 AGBW02012985 OWR44134.1 EAA44227.3 KA646264 AFP60893.1 CP012525 ALC44907.1 CH902618 EDV40438.1 CH933809 EDW18697.1 GDHF01006779 JAI45535.1 ALC44900.1 CM002912 KMZ00102.1 GFDL01004884 JAV30161.1 GGFL01006404 MBW70582.1 CM000159 EDW94801.1 EDW73064.2 AXCM01009805 CH480815 EDW41803.1 AE014296 AY069733 AAF49418.2 AAL39878.1 GAMC01007113 JAB99442.1 CH954178 EDV52042.2 GFDF01005306 JAV08778.1 CM000363 EDX10773.1 GGFM01006254 MBW27005.1 CH916366 EDV96393.1 JX460804 AFS49707.1 CH940647 EDW69361.1 GGFJ01004382 MBW53523.1 NEVH01021237 PNF19653.1 ETN64738.1 NEVH01024540 PNF16841.1 RSAL01000077 RVE48784.1 ATLV01010792 KE524614 KFB35691.1 KK853013 KDR12525.1 ETN64736.1 KK852732 KDR17469.1 GEDC01016220 JAS21078.1 GFDF01005304 JAV08780.1 GFDF01005305 JAV08779.1 JN033726 AEW46879.1 CH479217 EDW33737.1
AB264710 BAG30785.1 RSAL01000018 RVE52880.1 LN851681 CRL08768.1 NWSH01007170 PCG63058.1 KQ459606 KPI91416.1 PCG63057.1 KQ459984 KPJ18942.1 BABH01014157 NWSH01000598 PCG75431.1 KQ459589 KPI97521.1 LT635664 SGZ49394.1 ODYU01004836 SOQ45125.1 RSAL01000086 RVE48281.1 KQ460870 KPJ11681.1 DS232013 EDS31873.1 KT992464 AOY34571.1 CH963847 EDW73065.2 AXCN02000455 JXUM01031352 KQ560919 KXJ80348.1 GFDL01002519 JAV32526.1 CH477568 EAT38910.1 GBXI01000924 JAD13368.1 GGFL01002037 MBW66215.1 GBXI01006185 JAD08107.1 CH478582 EAT32750.1 GAKP01018616 JAC40336.1 LJIG01003002 KRT83938.1 GAKP01018617 JAC40335.1 JXJN01024604 JXJN01024605 JXJN01024606 GDHF01025685 JAI26629.1 ADMH02000892 ETN64734.1 AAAB01008960 EAL40075.2 APCN01002262 GDHF01006374 JAI45940.1 AGBW02012985 OWR44134.1 EAA44227.3 KA646264 AFP60893.1 CP012525 ALC44907.1 CH902618 EDV40438.1 CH933809 EDW18697.1 GDHF01006779 JAI45535.1 ALC44900.1 CM002912 KMZ00102.1 GFDL01004884 JAV30161.1 GGFL01006404 MBW70582.1 CM000159 EDW94801.1 EDW73064.2 AXCM01009805 CH480815 EDW41803.1 AE014296 AY069733 AAF49418.2 AAL39878.1 GAMC01007113 JAB99442.1 CH954178 EDV52042.2 GFDF01005306 JAV08778.1 CM000363 EDX10773.1 GGFM01006254 MBW27005.1 CH916366 EDV96393.1 JX460804 AFS49707.1 CH940647 EDW69361.1 GGFJ01004382 MBW53523.1 NEVH01021237 PNF19653.1 ETN64738.1 NEVH01024540 PNF16841.1 RSAL01000077 RVE48784.1 ATLV01010792 KE524614 KFB35691.1 KK853013 KDR12525.1 ETN64736.1 KK852732 KDR17469.1 GEDC01016220 JAS21078.1 GFDF01005304 JAV08780.1 GFDF01005305 JAV08779.1 JN033726 AEW46879.1 CH479217 EDW33737.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000075880 UP000002320 UP000076408 UP000007798 UP000075886 UP000069940 UP000249989 UP000091820 UP000075920 UP000008820 UP000192223 UP000078200 UP000092460 UP000192221 UP000092443 UP000092445 UP000000673 UP000007062 UP000075882 UP000075903 UP000075840 UP000075902 UP000092553 UP000007801 UP000009192 UP000002282 UP000075883 UP000001292 UP000000803 UP000076407 UP000008711 UP000000304 UP000001070 UP000008792 UP000235965 UP000095300 UP000030765 UP000027135 UP000008744
UP000075880 UP000002320 UP000076408 UP000007798 UP000075886 UP000069940 UP000249989 UP000091820 UP000075920 UP000008820 UP000192223 UP000078200 UP000092460 UP000192221 UP000092443 UP000092445 UP000000673 UP000007062 UP000075882 UP000075903 UP000075840 UP000075902 UP000092553 UP000007801 UP000009192 UP000002282 UP000075883 UP000001292 UP000000803 UP000076407 UP000008711 UP000000304 UP000001070 UP000008792 UP000235965 UP000095300 UP000030765 UP000027135 UP000008744
Pfam
PF00232 Glyco_hydro_1
Interpro
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
H9IYW5
G9F9I2
A0A212EKQ3
B2DBM6
A0A3S2TR57
A0A0K2YR16
+ More
A0A2A4IUI6 A0A194PDJ4 A0A2A4ISU7 A0A194RN51 H9IZH5 A0A2A4JTU1 A0A194PVV9 A0A1W1EGM5 A0A2H1VWC6 A0A3S2M0F9 A0A182J2M6 A0A194R738 B0WNN7 A0A219WGQ6 A0A182YP84 B4MM56 A0A182QNT7 A0A182HAK7 A0A1A9W4X6 A0A1Q3FY94 A0A1Y9IW73 Q16WF2 A0A1W4XAC4 A0A0A1XS57 A0A182QI13 A0A1S4G3W5 A0A2M4CLR8 A0A0A1XAG3 Q16ET6 A0A034VAL4 A0A1A9UP13 A0A0T6B9C6 A0A034VEY7 A0A1B0C2P0 A0A1W4UV58 A0A1A9XR01 A0A1A9ZLX0 A0A0K8UIW8 W5JPP3 Q5TQY8 A0A182LL18 A0A1Y9IRY8 A0A1Y9GKT0 A0A182U0B4 A0A0K8W4X1 A0A1Y9IVZ0 A0A212ERQ2 Q7PI98 T1PC49 A0A0M4F1P7 B3M4E4 B4KXZ5 A0A0K8W3Q7 A0A0M4F0Y9 A0A0J9RWT3 A0A1Q3FR87 A0A2M4CZ21 B4PKA3 A0A182TWE4 B4MM55 A0A182LWK9 A0A1Y9IW90 A0A1Y9GL87 B4HK62 Q9VV98 A0A1Y9J0J6 W8BL15 B3NDI0 A0A1L8DQN1 B4QMY1 A0A2M3ZET6 B4IXF6 J9Z300 B4LG53 A0A2M4BKN2 A0A2J7PTJ0 W5JN46 A0A1I8PM40 A0A1Y9IS75 A0A2J7PKH8 A0A3S2PE71 A0A084VCJ7 A0A182YP82 A0A067R142 W5JKR6 A0A067RC84 A0A182JLR1 A0A1B6D5W8 A0A1L8DR18 A0A1Y9IRG1 A0A1L8DQS5 G9F9I1 A0A1Y9J0G9 B4H7D1
A0A2A4IUI6 A0A194PDJ4 A0A2A4ISU7 A0A194RN51 H9IZH5 A0A2A4JTU1 A0A194PVV9 A0A1W1EGM5 A0A2H1VWC6 A0A3S2M0F9 A0A182J2M6 A0A194R738 B0WNN7 A0A219WGQ6 A0A182YP84 B4MM56 A0A182QNT7 A0A182HAK7 A0A1A9W4X6 A0A1Q3FY94 A0A1Y9IW73 Q16WF2 A0A1W4XAC4 A0A0A1XS57 A0A182QI13 A0A1S4G3W5 A0A2M4CLR8 A0A0A1XAG3 Q16ET6 A0A034VAL4 A0A1A9UP13 A0A0T6B9C6 A0A034VEY7 A0A1B0C2P0 A0A1W4UV58 A0A1A9XR01 A0A1A9ZLX0 A0A0K8UIW8 W5JPP3 Q5TQY8 A0A182LL18 A0A1Y9IRY8 A0A1Y9GKT0 A0A182U0B4 A0A0K8W4X1 A0A1Y9IVZ0 A0A212ERQ2 Q7PI98 T1PC49 A0A0M4F1P7 B3M4E4 B4KXZ5 A0A0K8W3Q7 A0A0M4F0Y9 A0A0J9RWT3 A0A1Q3FR87 A0A2M4CZ21 B4PKA3 A0A182TWE4 B4MM55 A0A182LWK9 A0A1Y9IW90 A0A1Y9GL87 B4HK62 Q9VV98 A0A1Y9J0J6 W8BL15 B3NDI0 A0A1L8DQN1 B4QMY1 A0A2M3ZET6 B4IXF6 J9Z300 B4LG53 A0A2M4BKN2 A0A2J7PTJ0 W5JN46 A0A1I8PM40 A0A1Y9IS75 A0A2J7PKH8 A0A3S2PE71 A0A084VCJ7 A0A182YP82 A0A067R142 W5JKR6 A0A067RC84 A0A182JLR1 A0A1B6D5W8 A0A1L8DR18 A0A1Y9IRG1 A0A1L8DQS5 G9F9I1 A0A1Y9J0G9 B4H7D1
PDB
3VII
E-value=1.65384e-125,
Score=1152
Ontologies
GO
PANTHER
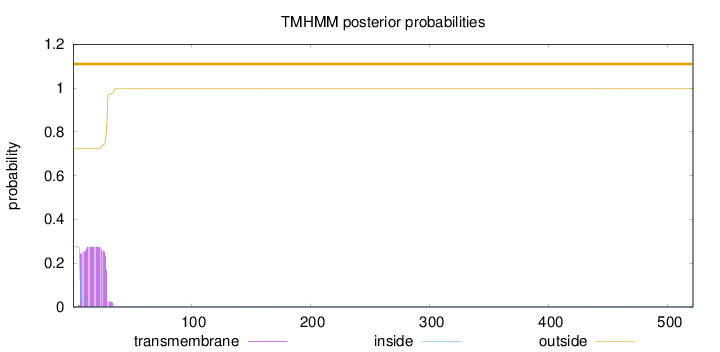
Topology
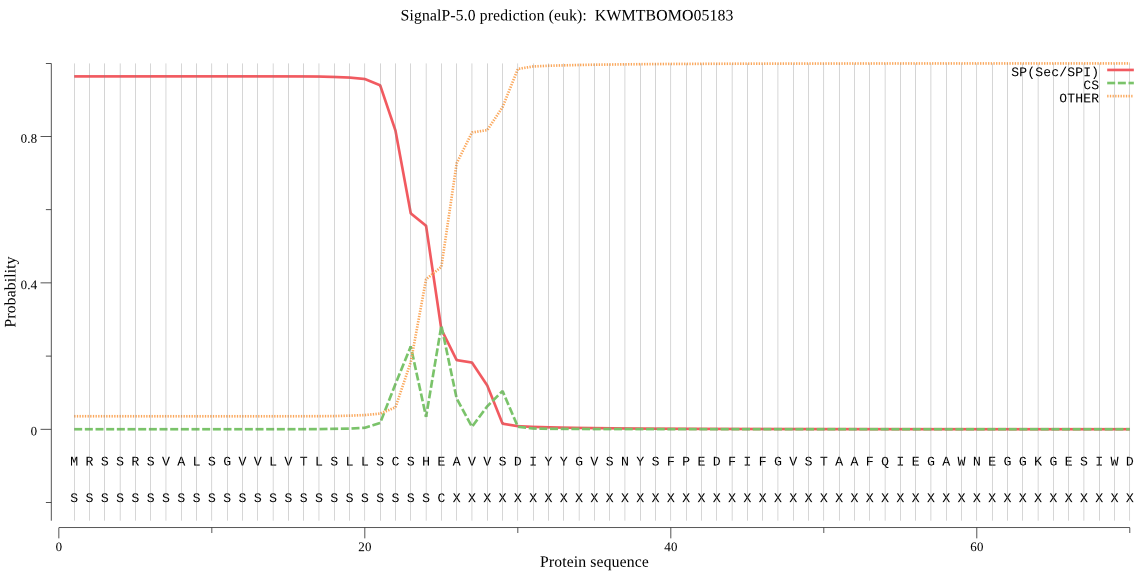
SignalP
Position: 1 - 25,
Likelihood: 0.964615
Length:
521
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.16178
Exp number, first 60 AAs:
6.1324
Total prob of N-in:
0.27740
outside
1 - 521
Population Genetic Test Statistics
Pi
205.168007
Theta
189.60417
Tajima's D
0.499611
CLR
0.415583
CSRT
0.513924303784811
Interpretation
Uncertain
