Gene
KWMTBOMO05178 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012485
Annotation
phosphoserine_aminotransferase_1_[Bombyx_mori]
Full name
Phosphoserine aminotransferase
+ More
Probable phosphoserine aminotransferase
Probable phosphoserine aminotransferase
Alternative Name
Phosphohydroxythreonine aminotransferase
Location in the cell
Cytoplasmic Reliability : 1.999
Sequence
CDS
ATGAAGCTAAATGTCGAAATTCAGGATGTTGTGCGAAACTTATTAGATGTACCGGACAACTACAAAGTGCTGTTTTTAGCTGGTGGGGGTCTTGGTCAATTTGCAGCTGTTCCTTTAAACTTAATATCCAGAACTGGTACTGCTGATTATGTAGTAACAGGTGCGTGGTCTGCTAAAGCAGCCAAGGAGGCGAAAAAATATGGGAAAGTAAATTTGGTGCTACCACCGACAGATAAATATGAAGACATACCTGATCAGACAAAATGGAACCTTGATCCCAATGCTTCATATGTACATATTTGTACTAATGAAACAATACATGGTGTTGAATTTGACTTCATACCAGATACGAAAGGAGTACCTTTAATCGCAGATATGTCCTCAAACATTATGTCGAAGAAAGTTGATGTTTCAAAGTTTGGGGTGATATATGCTGGTGCTCAAAAGAATATTGGTACATCTGGGGTAGCGCTTGTCATTGTTAGAGAGGATCTTTTGAATCAGGCTCTACCGACATGTCCCTCTTTATTAGACTGGACAGCTAACTACAAACAGAATTCAATTTTAAACACTCCACCTATGTTTGCCATCTATATAATGGGTCGAGTATTACAATGGATTCAAAGGAATGGAGGATTAGAAGGAATGTCTCAATTGGCAACAAAGAAAGCCTCACTTATCTACAACACAATTGAGCAATCAAATGGTTTTTATTATGCCCCTGTAGCTAAAAATGTTCGCAGTAAAATGAATGTGCCATTCAGAATTGGTTGCCCTGGTGATGATGCCTTAGAAAAAGAATTCTTGAAGGGTGCTGAGACTTTGGGACTTATTCAGCTAAAAGGACACAGAGACGTTGGCGGTATTCGTGCCTCAATATATAATGCAGTGACCCTAGAGGAGGTTCAAGCTCTCGTTCAGTACATGGAGGAGTTCTATAAAAAACATTCCAAGTAA
Protein
MKLNVEIQDVVRNLLDVPDNYKVLFLAGGGLGQFAAVPLNLISRTGTADYVVTGAWSAKAAKEAKKYGKVNLVLPPTDKYEDIPDQTKWNLDPNASYVHICTNETIHGVEFDFIPDTKGVPLIADMSSNIMSKKVDVSKFGVIYAGAQKNIGTSGVALVIVREDLLNQALPTCPSLLDWTANYKQNSILNTPPMFAIYIMGRVLQWIQRNGGLEGMSQLATKKASLIYNTIEQSNGFYYAPVAKNVRSKMNVPFRIGCPGDDALEKEFLKGAETLGLIQLKGHRDVGGIRASIYNAVTLEEVQALVQYMEEFYKKHSK
Summary
Description
Catalyzes the reversible conversion of 3-phosphohydroxypyruvate to phosphoserine and of 3-hydroxy-2-oxo-4-phosphonooxybutanoate to phosphohydroxythreonine.
Catalytic Activity
2-oxoglutarate + O-phospho-L-serine = 3-phosphooxypyruvate + L-glutamate
2-oxoglutarate + 4-(phosphooxy)-L-threonine = (R)-3-hydroxy-2-oxo-4-phosphooxybutanoate + L-glutamate
2-oxoglutarate + 4-(phosphooxy)-L-threonine = (R)-3-hydroxy-2-oxo-4-phosphooxybutanoate + L-glutamate
Cofactor
pyridoxal 5'-phosphate
Subunit
Homodimer.
Similarity
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family.
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family. SerC subfamily.
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family. SerC subfamily.
Keywords
Amino-acid biosynthesis
Aminotransferase
Complete proteome
Pyridoxal phosphate
Reference proteome
Serine biosynthesis
Transferase
Feature
chain Probable phosphoserine aminotransferase
Uniprot
Q2F5M8
H9JSH4
E3VVV6
A0A3G1T1N2
A0A2H1V2G6
A0A2A4J2R0
+ More
S4NNM6 I4DP14 A0A194PDS2 A0A194RM83 A0A067RGP0 A0A2P8XZ63 A0A068TK14 A0A2J7RRU7 K7J235 A0A182V703 A0A182XDN6 T1HR77 A0A182LNM1 Q5TRW7 A0A182IDE4 A0A0N7Z997 R4G458 A0A182YG81 A0A182PWA7 A0A0A9YMV0 E0VYZ3 A0A182J0L0 A0A182KCK7 A0A170Z554 E2BFQ7 A0A026WBG9 A0A084W7S2 A0A1L8DZL1 A0A336MJR0 A0A182VPN8 A0A182H1Q3 A0A182RED5 A0A1L8DZU0 Q3YMT5 Q9VAN0 A0A1B0AGQ0 B4GPH5 A0A182QK31 A0A1A9VKP0 B4PQ55 A0A2M4AE96 A0A2M4ADR6 A0A2M4AHT4 A0A2A3E3I4 A0A1A9Y8Z4 A0A1B0BGU3 A0A1A9WMI0 B4HZ76 Q1HRL1 A0A1B0CAY7 A0A0J7KU07 Q29BY2 D3TLH6 Q16LP7 B3P5J6 A0A1J1HLT9 B0WL19 A0A182FAJ4 A0A2M4BRY7 A0A2M3Z4S6 A0A088ADX3 E2AJ73 A0A182NN17 W5JTQ3 T1E7S4 A0A0A1WFM0 A0A1I8PD33 A0A2M4BRV3 A0A0L7RGE4 A0A3B0JLW8 U5EWV5 A0A1W4U5K1 A0A1L8EEQ1 A0A3B0K8H2 A0A1L8EEM4 A0A1Y1K9Q8 B4K5X3 A0A1L8EF11 A0A1Q3FLL2 A0A3B4C4Z9 B4NFM9 Q803I7 A0A0C9PTY7 A0A3N0YDT1 Q6P2U8 A0A0L0CEB1 A0A034WIQ8 W5LMK1 A0A151XDF9 A0A369SMH2 B3S7Q4 A0A3B4UL75 A0A3B4YCU3 A0A3B5AWW6
S4NNM6 I4DP14 A0A194PDS2 A0A194RM83 A0A067RGP0 A0A2P8XZ63 A0A068TK14 A0A2J7RRU7 K7J235 A0A182V703 A0A182XDN6 T1HR77 A0A182LNM1 Q5TRW7 A0A182IDE4 A0A0N7Z997 R4G458 A0A182YG81 A0A182PWA7 A0A0A9YMV0 E0VYZ3 A0A182J0L0 A0A182KCK7 A0A170Z554 E2BFQ7 A0A026WBG9 A0A084W7S2 A0A1L8DZL1 A0A336MJR0 A0A182VPN8 A0A182H1Q3 A0A182RED5 A0A1L8DZU0 Q3YMT5 Q9VAN0 A0A1B0AGQ0 B4GPH5 A0A182QK31 A0A1A9VKP0 B4PQ55 A0A2M4AE96 A0A2M4ADR6 A0A2M4AHT4 A0A2A3E3I4 A0A1A9Y8Z4 A0A1B0BGU3 A0A1A9WMI0 B4HZ76 Q1HRL1 A0A1B0CAY7 A0A0J7KU07 Q29BY2 D3TLH6 Q16LP7 B3P5J6 A0A1J1HLT9 B0WL19 A0A182FAJ4 A0A2M4BRY7 A0A2M3Z4S6 A0A088ADX3 E2AJ73 A0A182NN17 W5JTQ3 T1E7S4 A0A0A1WFM0 A0A1I8PD33 A0A2M4BRV3 A0A0L7RGE4 A0A3B0JLW8 U5EWV5 A0A1W4U5K1 A0A1L8EEQ1 A0A3B0K8H2 A0A1L8EEM4 A0A1Y1K9Q8 B4K5X3 A0A1L8EF11 A0A1Q3FLL2 A0A3B4C4Z9 B4NFM9 Q803I7 A0A0C9PTY7 A0A3N0YDT1 Q6P2U8 A0A0L0CEB1 A0A034WIQ8 W5LMK1 A0A151XDF9 A0A369SMH2 B3S7Q4 A0A3B4UL75 A0A3B4YCU3 A0A3B5AWW6
EC Number
2.6.1.52
Pubmed
19121390
23622113
22651552
26354079
24845553
29403074
+ More
20075255 20966253 12364791 14747013 17210077 27129103 25244985 25401762 26823975 20566863 20798317 24508170 30249741 24438588 26483478 15917496 17994087 10731132 12537572 12537569 17550304 17204158 15632085 20353571 17510324 20920257 23761445 25830018 28004739 23594743 26108605 25348373 25329095 30042472 18719581
20075255 20966253 12364791 14747013 17210077 27129103 25244985 25401762 26823975 20566863 20798317 24508170 30249741 24438588 26483478 15917496 17994087 10731132 12537572 12537569 17550304 17204158 15632085 20353571 17510324 20920257 23761445 25830018 28004739 23594743 26108605 25348373 25329095 30042472 18719581
EMBL
DQ311395
KM027247
ABD36339.1
AIU94616.1
BABH01027841
HQ260728
+ More
ADO79970.1 MG992460 AXY94898.1 ODYU01000357 SOQ34999.1 NWSH01003803 PCG65804.1 GAIX01013866 JAA78694.1 AK403243 BAM19654.1 KQ459606 KPI91407.1 KQ459984 KPJ18933.1 KK852651 KDR19432.1 PYGN01001136 PSN37290.1 HG965794 CDO39390.1 NEVH01000598 PNF43557.1 ACPB03014376 AAAB01008952 EAL40174.2 APCN01005968 GDKW01001155 JAI55440.1 GAHY01001076 JAA76434.1 GBHO01010653 GBRD01018138 GDHC01011487 JAG32951.1 JAG47689.1 JAQ07142.1 DS235848 EEB18599.1 GEMB01002592 JAS00597.1 GL448039 EFN85445.1 KK107293 QOIP01000005 EZA53405.1 RLU23179.1 ATLV01021293 KE525316 KFB46266.1 GFDF01002176 JAV11908.1 UFQT01001354 SSX30170.1 JXUM01023194 KQ560654 KXJ81423.1 GFDF01002175 JAV11909.1 DQ062791 CM000364 AAY56664.1 EDX14821.1 DQ062790 AE014297 AY119560 CH479186 EDW39058.1 AXCN02001593 CM000160 EDW98324.1 GGFK01005607 MBW38928.1 GGFK01005606 MBW38927.1 GGFK01007013 MBW40334.1 KZ288416 PBC26044.1 JXJN01013988 CH480819 EDW53333.1 DQ440083 ABF18116.1 AJWK01004661 LBMM01003236 KMQ93776.1 CM000070 EAL26864.3 CCAG010023851 EZ422278 ADD18554.1 CH477898 EAT35240.1 CH954182 EDV53246.1 CVRI01000004 CRK87193.1 DS231979 EDS30182.1 GGFJ01006679 MBW55820.1 GGFM01002752 MBW23503.1 GL439967 EFN66513.1 ADMH02000475 ETN66335.1 GAMD01002797 JAA98793.1 GBXI01017084 JAC97207.1 GGFJ01006676 MBW55817.1 KQ414596 KOC69904.1 OUUW01000005 SPP81322.1 GANO01001249 JAB58622.1 GFDG01001621 JAV17178.1 SPP81321.1 GFDG01001622 JAV17177.1 GEZM01093706 JAV56176.1 CH933806 EDW16210.1 GFDG01001620 JAV17179.1 GFDL01006546 JAV28499.1 CH964251 EDW83096.1 LO017803 BC044467 AAH44467.1 GBYB01004778 JAG74545.1 RJVU01046259 ROL44389.1 BC064289 AAH64289.1 JRES01000501 KNC30748.1 GAKP01004383 JAC54569.1 KQ982294 KYQ58397.1 NOWV01000002 RDD47614.1 DS985254 EDV21334.1
ADO79970.1 MG992460 AXY94898.1 ODYU01000357 SOQ34999.1 NWSH01003803 PCG65804.1 GAIX01013866 JAA78694.1 AK403243 BAM19654.1 KQ459606 KPI91407.1 KQ459984 KPJ18933.1 KK852651 KDR19432.1 PYGN01001136 PSN37290.1 HG965794 CDO39390.1 NEVH01000598 PNF43557.1 ACPB03014376 AAAB01008952 EAL40174.2 APCN01005968 GDKW01001155 JAI55440.1 GAHY01001076 JAA76434.1 GBHO01010653 GBRD01018138 GDHC01011487 JAG32951.1 JAG47689.1 JAQ07142.1 DS235848 EEB18599.1 GEMB01002592 JAS00597.1 GL448039 EFN85445.1 KK107293 QOIP01000005 EZA53405.1 RLU23179.1 ATLV01021293 KE525316 KFB46266.1 GFDF01002176 JAV11908.1 UFQT01001354 SSX30170.1 JXUM01023194 KQ560654 KXJ81423.1 GFDF01002175 JAV11909.1 DQ062791 CM000364 AAY56664.1 EDX14821.1 DQ062790 AE014297 AY119560 CH479186 EDW39058.1 AXCN02001593 CM000160 EDW98324.1 GGFK01005607 MBW38928.1 GGFK01005606 MBW38927.1 GGFK01007013 MBW40334.1 KZ288416 PBC26044.1 JXJN01013988 CH480819 EDW53333.1 DQ440083 ABF18116.1 AJWK01004661 LBMM01003236 KMQ93776.1 CM000070 EAL26864.3 CCAG010023851 EZ422278 ADD18554.1 CH477898 EAT35240.1 CH954182 EDV53246.1 CVRI01000004 CRK87193.1 DS231979 EDS30182.1 GGFJ01006679 MBW55820.1 GGFM01002752 MBW23503.1 GL439967 EFN66513.1 ADMH02000475 ETN66335.1 GAMD01002797 JAA98793.1 GBXI01017084 JAC97207.1 GGFJ01006676 MBW55817.1 KQ414596 KOC69904.1 OUUW01000005 SPP81322.1 GANO01001249 JAB58622.1 GFDG01001621 JAV17178.1 SPP81321.1 GFDG01001622 JAV17177.1 GEZM01093706 JAV56176.1 CH933806 EDW16210.1 GFDG01001620 JAV17179.1 GFDL01006546 JAV28499.1 CH964251 EDW83096.1 LO017803 BC044467 AAH44467.1 GBYB01004778 JAG74545.1 RJVU01046259 ROL44389.1 BC064289 AAH64289.1 JRES01000501 KNC30748.1 GAKP01004383 JAC54569.1 KQ982294 KYQ58397.1 NOWV01000002 RDD47614.1 DS985254 EDV21334.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000027135
UP000245037
+ More
UP000235965 UP000002358 UP000075903 UP000076407 UP000015103 UP000075882 UP000007062 UP000075840 UP000076408 UP000075885 UP000009046 UP000075880 UP000075881 UP000008237 UP000053097 UP000279307 UP000030765 UP000075920 UP000069940 UP000249989 UP000075900 UP000000304 UP000000803 UP000092445 UP000008744 UP000075886 UP000078200 UP000002282 UP000242457 UP000092443 UP000092460 UP000091820 UP000001292 UP000092461 UP000036403 UP000001819 UP000092444 UP000008820 UP000008711 UP000183832 UP000002320 UP000069272 UP000005203 UP000000311 UP000075884 UP000000673 UP000095300 UP000053825 UP000268350 UP000192221 UP000009192 UP000261440 UP000007798 UP000000437 UP000037069 UP000018467 UP000075809 UP000253843 UP000009022 UP000261420 UP000261360 UP000261400
UP000235965 UP000002358 UP000075903 UP000076407 UP000015103 UP000075882 UP000007062 UP000075840 UP000076408 UP000075885 UP000009046 UP000075880 UP000075881 UP000008237 UP000053097 UP000279307 UP000030765 UP000075920 UP000069940 UP000249989 UP000075900 UP000000304 UP000000803 UP000092445 UP000008744 UP000075886 UP000078200 UP000002282 UP000242457 UP000092443 UP000092460 UP000091820 UP000001292 UP000092461 UP000036403 UP000001819 UP000092444 UP000008820 UP000008711 UP000183832 UP000002320 UP000069272 UP000005203 UP000000311 UP000075884 UP000000673 UP000095300 UP000053825 UP000268350 UP000192221 UP000009192 UP000261440 UP000007798 UP000000437 UP000037069 UP000018467 UP000075809 UP000253843 UP000009022 UP000261420 UP000261360 UP000261400
Interpro
IPR020578
Aminotrans_V_PyrdxlP_BS
+ More
IPR015421 PyrdxlP-dep_Trfase_major
IPR000192 Aminotrans_V_dom
IPR015424 PyrdxlP-dep_Trfase
IPR022278 Pser_aminoTfrase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR013288 Cyt_c_oxidase_su4
IPR036639 Cyt_c_oxidase_su4_sf
IPR004203 Cyt_c_oxidase_su4_fam
IPR007110 Ig-like_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR000192 Aminotrans_V_dom
IPR015424 PyrdxlP-dep_Trfase
IPR022278 Pser_aminoTfrase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR013288 Cyt_c_oxidase_su4
IPR036639 Cyt_c_oxidase_su4_sf
IPR004203 Cyt_c_oxidase_su4_fam
IPR007110 Ig-like_dom
Gene 3D
ProteinModelPortal
Q2F5M8
H9JSH4
E3VVV6
A0A3G1T1N2
A0A2H1V2G6
A0A2A4J2R0
+ More
S4NNM6 I4DP14 A0A194PDS2 A0A194RM83 A0A067RGP0 A0A2P8XZ63 A0A068TK14 A0A2J7RRU7 K7J235 A0A182V703 A0A182XDN6 T1HR77 A0A182LNM1 Q5TRW7 A0A182IDE4 A0A0N7Z997 R4G458 A0A182YG81 A0A182PWA7 A0A0A9YMV0 E0VYZ3 A0A182J0L0 A0A182KCK7 A0A170Z554 E2BFQ7 A0A026WBG9 A0A084W7S2 A0A1L8DZL1 A0A336MJR0 A0A182VPN8 A0A182H1Q3 A0A182RED5 A0A1L8DZU0 Q3YMT5 Q9VAN0 A0A1B0AGQ0 B4GPH5 A0A182QK31 A0A1A9VKP0 B4PQ55 A0A2M4AE96 A0A2M4ADR6 A0A2M4AHT4 A0A2A3E3I4 A0A1A9Y8Z4 A0A1B0BGU3 A0A1A9WMI0 B4HZ76 Q1HRL1 A0A1B0CAY7 A0A0J7KU07 Q29BY2 D3TLH6 Q16LP7 B3P5J6 A0A1J1HLT9 B0WL19 A0A182FAJ4 A0A2M4BRY7 A0A2M3Z4S6 A0A088ADX3 E2AJ73 A0A182NN17 W5JTQ3 T1E7S4 A0A0A1WFM0 A0A1I8PD33 A0A2M4BRV3 A0A0L7RGE4 A0A3B0JLW8 U5EWV5 A0A1W4U5K1 A0A1L8EEQ1 A0A3B0K8H2 A0A1L8EEM4 A0A1Y1K9Q8 B4K5X3 A0A1L8EF11 A0A1Q3FLL2 A0A3B4C4Z9 B4NFM9 Q803I7 A0A0C9PTY7 A0A3N0YDT1 Q6P2U8 A0A0L0CEB1 A0A034WIQ8 W5LMK1 A0A151XDF9 A0A369SMH2 B3S7Q4 A0A3B4UL75 A0A3B4YCU3 A0A3B5AWW6
S4NNM6 I4DP14 A0A194PDS2 A0A194RM83 A0A067RGP0 A0A2P8XZ63 A0A068TK14 A0A2J7RRU7 K7J235 A0A182V703 A0A182XDN6 T1HR77 A0A182LNM1 Q5TRW7 A0A182IDE4 A0A0N7Z997 R4G458 A0A182YG81 A0A182PWA7 A0A0A9YMV0 E0VYZ3 A0A182J0L0 A0A182KCK7 A0A170Z554 E2BFQ7 A0A026WBG9 A0A084W7S2 A0A1L8DZL1 A0A336MJR0 A0A182VPN8 A0A182H1Q3 A0A182RED5 A0A1L8DZU0 Q3YMT5 Q9VAN0 A0A1B0AGQ0 B4GPH5 A0A182QK31 A0A1A9VKP0 B4PQ55 A0A2M4AE96 A0A2M4ADR6 A0A2M4AHT4 A0A2A3E3I4 A0A1A9Y8Z4 A0A1B0BGU3 A0A1A9WMI0 B4HZ76 Q1HRL1 A0A1B0CAY7 A0A0J7KU07 Q29BY2 D3TLH6 Q16LP7 B3P5J6 A0A1J1HLT9 B0WL19 A0A182FAJ4 A0A2M4BRY7 A0A2M3Z4S6 A0A088ADX3 E2AJ73 A0A182NN17 W5JTQ3 T1E7S4 A0A0A1WFM0 A0A1I8PD33 A0A2M4BRV3 A0A0L7RGE4 A0A3B0JLW8 U5EWV5 A0A1W4U5K1 A0A1L8EEQ1 A0A3B0K8H2 A0A1L8EEM4 A0A1Y1K9Q8 B4K5X3 A0A1L8EF11 A0A1Q3FLL2 A0A3B4C4Z9 B4NFM9 Q803I7 A0A0C9PTY7 A0A3N0YDT1 Q6P2U8 A0A0L0CEB1 A0A034WIQ8 W5LMK1 A0A151XDF9 A0A369SMH2 B3S7Q4 A0A3B4UL75 A0A3B4YCU3 A0A3B5AWW6
PDB
3E77
E-value=4.3765e-97,
Score=904
Ontologies
PATHWAY
00260
Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00750 Vitamin B6 metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00750 Vitamin B6 metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
Topology
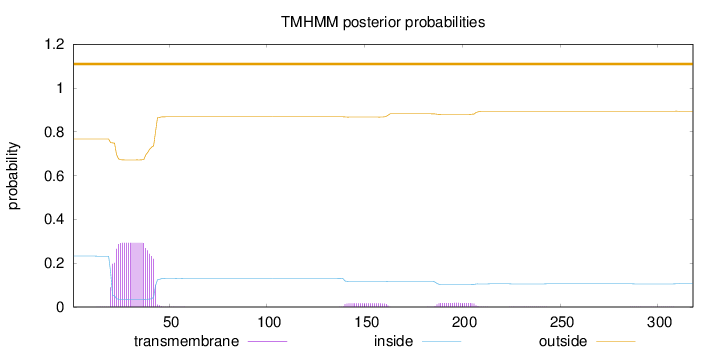
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.94478
Exp number, first 60 AAs:
6.20433
Total prob of N-in:
0.23247
outside
1 - 318
Population Genetic Test Statistics
Pi
198.30892
Theta
154.906934
Tajima's D
0.403653
CLR
0.422664
CSRT
0.485575721213939
Interpretation
Uncertain
