Gene
KWMTBOMO05176
Pre Gene Modal
BGIBMGA012489
Annotation
PREDICTED:_coronin-2B-like_isoform_X1_[Bombyx_mori]
Full name
Coronin
Location in the cell
Nuclear Reliability : 2.688
Sequence
CDS
ATGTTCCTCTCGCGCGAAACCACACCCGACTACCGCCCTCTCAACGCCTGCCCGAACGCACTCAATCACAACAATGAAAATGAGATACAAGTGAACGAGAAATCGCAAAAGACGAACGCCAATCAGAACACGAAGTTCCATCAGCTTCAACGCATGTTCGGCAAGCAGACGAACGACGCGGAGCCGGTGCCCCTCTACAAGCAGATCAACCAGGGAGACGTCTTCAGCACCGAGCACGAGCTACGCCTGGCCTTCAATCGTCAAGGAGAAGAACTCAGGATCGTAAAGAGGCAGCTTCAGAACAGTCAGCAGAGAGTGCGGGAGCTGGAACAGCACGTCGCCGCTCTGCAAGCCAAGCTGCAGTGA
Protein
MFLSRETTPDYRPLNACPNALNHNNENEIQVNEKSQKTNANQNTKFHQLQRMFGKQTNDAEPVPLYKQINQGDVFSTEHELRLAFNRQGEELRIVKRQLQNSQQRVRELEQHVAALQAKLQ
Summary
Similarity
Belongs to the WD repeat coronin family.
Feature
chain Coronin
Uniprot
H9JSH8
A0A2H1VCY9
A0A3S2NN17
A0A194PDI4
A0A0L7KVW0
A0A194RS87
+ More
A0A154PC19 A0A2A3EMU2 A0A088ADK7 A0A0L7RH42 A0A3L8DT04 A0A026VYD0 A0A232FAM4 E1ZVU7 E2BW29 K7IZY9 A0A1E1W3P5 A0A0M8ZR10 E9JD38 A0A2P8YYH3 A0A1B6L791 A0A1B6ES35 A0A195FWZ9 F4WLT3 A0A158NXT0 A0A151I4X2 A0A0J7NLY3 A0A1B6E255 A0A1B6ICR8 Q17LA0 D7EIA7 A0A2S2NN35 B0X5D8 A0A1Q3FIR5 A0A1Q3FIV4 A0A1Q3FIT3 A0A1Q3FIW4 A0A1Q3FIM7 A0A1Q3FIN1 A0A182NHR7 A0A084WNF4 A0A182WJ48 W5JI75 A0A182FF80 A0A2H8TIU0 A0A182VE52 A0A182X3M1 A0A182TZE3 Q5TWZ8 A0A182Q9R9 A0A182ING4 A0A182I3E8 J9JPP8 A0A182PQB3 A0A182M4M3 A0A182R4D7 A0A1S4EYR7 A0A182XYZ9 A0A3Q0J4K4 A0A182KRH1 A0A182K7D6 A0A1W4X7R0 A0A1W4X7J6 A0A1Y1LHM6 N6T524 A0A182S6W8 T1HSS1 A0A2J7PT74 A0A336MVG0 A0A0V0GCB5 A0A067QVB2 A0A069DVQ5 A0A023F3A1 A0A224XAA7 A0A0T6B5F8 A0A2J7PT65 A0A0P4VP43 A0A0K8T9B7 A0A0A9WTY6 A0A0A9XNQ9 A0A0A9Z3I3 U4U2X8 A0A0K8T8M4 A0A0A9Z5P6 A0A0A9WYP5 A0A0A9X2A0 A0A0A9XVE3 A0A0A9Z3H8 A0A0A9WEV6 T1JFZ6 A0A1J1HXI0 A0A0P4W5C1
A0A154PC19 A0A2A3EMU2 A0A088ADK7 A0A0L7RH42 A0A3L8DT04 A0A026VYD0 A0A232FAM4 E1ZVU7 E2BW29 K7IZY9 A0A1E1W3P5 A0A0M8ZR10 E9JD38 A0A2P8YYH3 A0A1B6L791 A0A1B6ES35 A0A195FWZ9 F4WLT3 A0A158NXT0 A0A151I4X2 A0A0J7NLY3 A0A1B6E255 A0A1B6ICR8 Q17LA0 D7EIA7 A0A2S2NN35 B0X5D8 A0A1Q3FIR5 A0A1Q3FIV4 A0A1Q3FIT3 A0A1Q3FIW4 A0A1Q3FIM7 A0A1Q3FIN1 A0A182NHR7 A0A084WNF4 A0A182WJ48 W5JI75 A0A182FF80 A0A2H8TIU0 A0A182VE52 A0A182X3M1 A0A182TZE3 Q5TWZ8 A0A182Q9R9 A0A182ING4 A0A182I3E8 J9JPP8 A0A182PQB3 A0A182M4M3 A0A182R4D7 A0A1S4EYR7 A0A182XYZ9 A0A3Q0J4K4 A0A182KRH1 A0A182K7D6 A0A1W4X7R0 A0A1W4X7J6 A0A1Y1LHM6 N6T524 A0A182S6W8 T1HSS1 A0A2J7PT74 A0A336MVG0 A0A0V0GCB5 A0A067QVB2 A0A069DVQ5 A0A023F3A1 A0A224XAA7 A0A0T6B5F8 A0A2J7PT65 A0A0P4VP43 A0A0K8T9B7 A0A0A9WTY6 A0A0A9XNQ9 A0A0A9Z3I3 U4U2X8 A0A0K8T8M4 A0A0A9Z5P6 A0A0A9WYP5 A0A0A9X2A0 A0A0A9XVE3 A0A0A9Z3H8 A0A0A9WEV6 T1JFZ6 A0A1J1HXI0 A0A0P4W5C1
Pubmed
EMBL
BABH01027842
BABH01027843
BABH01027844
BABH01027845
ODYU01001862
SOQ38656.1
+ More
RSAL01000028 RVE51831.1 KQ459606 KPI91406.1 JTDY01005097 KOB67373.1 KQ459984 KPJ18931.1 KQ434864 KZC08974.1 KZ288206 PBC33075.1 KQ414596 KOC70021.1 QOIP01000004 RLU23560.1 KK107579 EZA48650.1 NNAY01000597 OXU27488.1 GL434640 EFN74698.1 GL451091 EFN80061.1 GDQN01009470 JAT81584.1 KQ435944 KOX68216.1 GL771866 EFZ09319.1 PYGN01000285 PSN49299.1 GEBQ01020379 JAT19598.1 GECZ01029038 JAS40731.1 KQ981204 KYN44963.1 GL888216 EGI64815.1 ADTU01003503 ADTU01003504 ADTU01003505 ADTU01003506 KQ976462 KYM84653.1 LBMM01003474 KMQ93490.1 GEDC01005283 JAS32015.1 GECU01027764 GECU01022978 JAS79942.1 JAS84728.1 CH477217 EAT47475.1 KQ972797 EFA11755.2 GGMR01005945 MBY18564.1 DS232375 EDS40823.1 GFDL01007619 JAV27426.1 GFDL01007632 JAV27413.1 GFDL01007652 JAV27393.1 GFDL01007622 JAV27423.1 GFDL01007618 JAV27427.1 GFDL01007626 JAV27419.1 ATLV01024597 KE525352 KFB51748.1 ADMH02001422 ETN62600.1 GFXV01002155 MBW13960.1 AAAB01008807 EAL41772.2 AXCN02000376 AXCN02000377 APCN01000207 ABLF02030810 AXCM01003014 GEZM01055115 GEZM01055114 GEZM01055113 GEZM01055112 GEZM01055111 GEZM01055110 JAV73133.1 APGK01043770 KB741017 ENN75269.1 ACPB03015723 NEVH01021919 PNF19526.1 UFQT01003111 SSX34594.1 GECL01000655 JAP05469.1 KK852901 KDR14123.1 GBGD01001003 JAC87886.1 GBBI01002822 JAC15890.1 GFTR01007171 JAW09255.1 LJIG01009685 KRT82585.1 PNF19528.1 GDKW01003680 JAI52915.1 GBRD01004071 JAG61750.1 GBHO01032390 GBHO01031065 JAG11214.1 JAG12539.1 GBHO01044328 GBHO01031059 GBHO01022333 GBHO01003788 GBRD01004069 JAF99275.1 JAG12545.1 JAG21271.1 JAG39816.1 JAG61752.1 GBHO01031062 GBHO01003782 JAG12542.1 JAG39822.1 KB631644 ERL84951.1 GBRD01004070 JAG61751.1 GBHO01044325 GBHO01044319 GBHO01032392 GBHO01003785 GBRD01004067 JAF99278.1 JAF99284.1 JAG11212.1 JAG39819.1 JAG61754.1 GBHO01032391 GBHO01031063 JAG11213.1 JAG12541.1 GBHO01032389 GBHO01003781 JAG11215.1 JAG39823.1 GBHO01031066 GBHO01022374 JAG12538.1 JAG21230.1 GBHO01031064 GBHO01003787 JAG12540.1 JAG39817.1 GBHO01044327 GBHO01037290 JAF99276.1 JAG06314.1 JH432192 CVRI01000035 CRK92733.1 GDRN01071065 JAI63761.1
RSAL01000028 RVE51831.1 KQ459606 KPI91406.1 JTDY01005097 KOB67373.1 KQ459984 KPJ18931.1 KQ434864 KZC08974.1 KZ288206 PBC33075.1 KQ414596 KOC70021.1 QOIP01000004 RLU23560.1 KK107579 EZA48650.1 NNAY01000597 OXU27488.1 GL434640 EFN74698.1 GL451091 EFN80061.1 GDQN01009470 JAT81584.1 KQ435944 KOX68216.1 GL771866 EFZ09319.1 PYGN01000285 PSN49299.1 GEBQ01020379 JAT19598.1 GECZ01029038 JAS40731.1 KQ981204 KYN44963.1 GL888216 EGI64815.1 ADTU01003503 ADTU01003504 ADTU01003505 ADTU01003506 KQ976462 KYM84653.1 LBMM01003474 KMQ93490.1 GEDC01005283 JAS32015.1 GECU01027764 GECU01022978 JAS79942.1 JAS84728.1 CH477217 EAT47475.1 KQ972797 EFA11755.2 GGMR01005945 MBY18564.1 DS232375 EDS40823.1 GFDL01007619 JAV27426.1 GFDL01007632 JAV27413.1 GFDL01007652 JAV27393.1 GFDL01007622 JAV27423.1 GFDL01007618 JAV27427.1 GFDL01007626 JAV27419.1 ATLV01024597 KE525352 KFB51748.1 ADMH02001422 ETN62600.1 GFXV01002155 MBW13960.1 AAAB01008807 EAL41772.2 AXCN02000376 AXCN02000377 APCN01000207 ABLF02030810 AXCM01003014 GEZM01055115 GEZM01055114 GEZM01055113 GEZM01055112 GEZM01055111 GEZM01055110 JAV73133.1 APGK01043770 KB741017 ENN75269.1 ACPB03015723 NEVH01021919 PNF19526.1 UFQT01003111 SSX34594.1 GECL01000655 JAP05469.1 KK852901 KDR14123.1 GBGD01001003 JAC87886.1 GBBI01002822 JAC15890.1 GFTR01007171 JAW09255.1 LJIG01009685 KRT82585.1 PNF19528.1 GDKW01003680 JAI52915.1 GBRD01004071 JAG61750.1 GBHO01032390 GBHO01031065 JAG11214.1 JAG12539.1 GBHO01044328 GBHO01031059 GBHO01022333 GBHO01003788 GBRD01004069 JAF99275.1 JAG12545.1 JAG21271.1 JAG39816.1 JAG61752.1 GBHO01031062 GBHO01003782 JAG12542.1 JAG39822.1 KB631644 ERL84951.1 GBRD01004070 JAG61751.1 GBHO01044325 GBHO01044319 GBHO01032392 GBHO01003785 GBRD01004067 JAF99278.1 JAF99284.1 JAG11212.1 JAG39819.1 JAG61754.1 GBHO01032391 GBHO01031063 JAG11213.1 JAG12541.1 GBHO01032389 GBHO01003781 JAG11215.1 JAG39823.1 GBHO01031066 GBHO01022374 JAG12538.1 JAG21230.1 GBHO01031064 GBHO01003787 JAG12540.1 JAG39817.1 GBHO01044327 GBHO01037290 JAF99276.1 JAG06314.1 JH432192 CVRI01000035 CRK92733.1 GDRN01071065 JAI63761.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000037510
UP000053240
UP000076502
+ More
UP000242457 UP000005203 UP000053825 UP000279307 UP000053097 UP000215335 UP000000311 UP000008237 UP000002358 UP000053105 UP000245037 UP000078541 UP000007755 UP000005205 UP000078540 UP000036403 UP000008820 UP000007266 UP000002320 UP000075884 UP000030765 UP000075920 UP000000673 UP000069272 UP000075903 UP000076407 UP000075902 UP000007062 UP000075886 UP000075880 UP000075840 UP000007819 UP000075885 UP000075883 UP000075900 UP000076408 UP000079169 UP000075882 UP000075881 UP000192223 UP000019118 UP000075901 UP000015103 UP000235965 UP000027135 UP000030742 UP000183832
UP000242457 UP000005203 UP000053825 UP000279307 UP000053097 UP000215335 UP000000311 UP000008237 UP000002358 UP000053105 UP000245037 UP000078541 UP000007755 UP000005205 UP000078540 UP000036403 UP000008820 UP000007266 UP000002320 UP000075884 UP000030765 UP000075920 UP000000673 UP000069272 UP000075903 UP000076407 UP000075902 UP000007062 UP000075886 UP000075880 UP000075840 UP000007819 UP000075885 UP000075883 UP000075900 UP000076408 UP000079169 UP000075882 UP000075881 UP000192223 UP000019118 UP000075901 UP000015103 UP000235965 UP000027135 UP000030742 UP000183832
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JSH8
A0A2H1VCY9
A0A3S2NN17
A0A194PDI4
A0A0L7KVW0
A0A194RS87
+ More
A0A154PC19 A0A2A3EMU2 A0A088ADK7 A0A0L7RH42 A0A3L8DT04 A0A026VYD0 A0A232FAM4 E1ZVU7 E2BW29 K7IZY9 A0A1E1W3P5 A0A0M8ZR10 E9JD38 A0A2P8YYH3 A0A1B6L791 A0A1B6ES35 A0A195FWZ9 F4WLT3 A0A158NXT0 A0A151I4X2 A0A0J7NLY3 A0A1B6E255 A0A1B6ICR8 Q17LA0 D7EIA7 A0A2S2NN35 B0X5D8 A0A1Q3FIR5 A0A1Q3FIV4 A0A1Q3FIT3 A0A1Q3FIW4 A0A1Q3FIM7 A0A1Q3FIN1 A0A182NHR7 A0A084WNF4 A0A182WJ48 W5JI75 A0A182FF80 A0A2H8TIU0 A0A182VE52 A0A182X3M1 A0A182TZE3 Q5TWZ8 A0A182Q9R9 A0A182ING4 A0A182I3E8 J9JPP8 A0A182PQB3 A0A182M4M3 A0A182R4D7 A0A1S4EYR7 A0A182XYZ9 A0A3Q0J4K4 A0A182KRH1 A0A182K7D6 A0A1W4X7R0 A0A1W4X7J6 A0A1Y1LHM6 N6T524 A0A182S6W8 T1HSS1 A0A2J7PT74 A0A336MVG0 A0A0V0GCB5 A0A067QVB2 A0A069DVQ5 A0A023F3A1 A0A224XAA7 A0A0T6B5F8 A0A2J7PT65 A0A0P4VP43 A0A0K8T9B7 A0A0A9WTY6 A0A0A9XNQ9 A0A0A9Z3I3 U4U2X8 A0A0K8T8M4 A0A0A9Z5P6 A0A0A9WYP5 A0A0A9X2A0 A0A0A9XVE3 A0A0A9Z3H8 A0A0A9WEV6 T1JFZ6 A0A1J1HXI0 A0A0P4W5C1
A0A154PC19 A0A2A3EMU2 A0A088ADK7 A0A0L7RH42 A0A3L8DT04 A0A026VYD0 A0A232FAM4 E1ZVU7 E2BW29 K7IZY9 A0A1E1W3P5 A0A0M8ZR10 E9JD38 A0A2P8YYH3 A0A1B6L791 A0A1B6ES35 A0A195FWZ9 F4WLT3 A0A158NXT0 A0A151I4X2 A0A0J7NLY3 A0A1B6E255 A0A1B6ICR8 Q17LA0 D7EIA7 A0A2S2NN35 B0X5D8 A0A1Q3FIR5 A0A1Q3FIV4 A0A1Q3FIT3 A0A1Q3FIW4 A0A1Q3FIM7 A0A1Q3FIN1 A0A182NHR7 A0A084WNF4 A0A182WJ48 W5JI75 A0A182FF80 A0A2H8TIU0 A0A182VE52 A0A182X3M1 A0A182TZE3 Q5TWZ8 A0A182Q9R9 A0A182ING4 A0A182I3E8 J9JPP8 A0A182PQB3 A0A182M4M3 A0A182R4D7 A0A1S4EYR7 A0A182XYZ9 A0A3Q0J4K4 A0A182KRH1 A0A182K7D6 A0A1W4X7R0 A0A1W4X7J6 A0A1Y1LHM6 N6T524 A0A182S6W8 T1HSS1 A0A2J7PT74 A0A336MVG0 A0A0V0GCB5 A0A067QVB2 A0A069DVQ5 A0A023F3A1 A0A224XAA7 A0A0T6B5F8 A0A2J7PT65 A0A0P4VP43 A0A0K8T9B7 A0A0A9WTY6 A0A0A9XNQ9 A0A0A9Z3I3 U4U2X8 A0A0K8T8M4 A0A0A9Z5P6 A0A0A9WYP5 A0A0A9X2A0 A0A0A9XVE3 A0A0A9Z3H8 A0A0A9WEV6 T1JFZ6 A0A1J1HXI0 A0A0P4W5C1
Ontologies
PANTHER
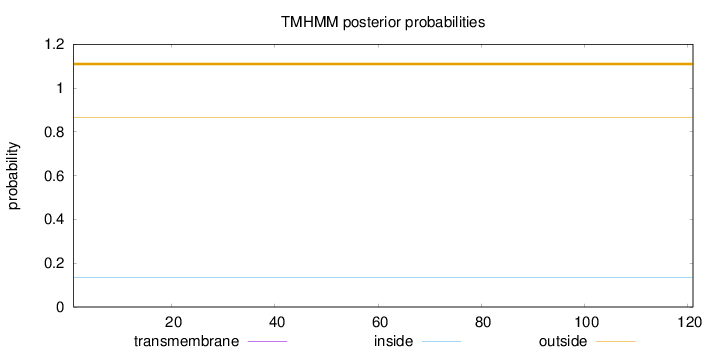
Topology
Length:
121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13362
outside
1 - 121
Population Genetic Test Statistics
Pi
235.671604
Theta
176.153641
Tajima's D
1.110224
CLR
0.00855
CSRT
0.691465426728664
Interpretation
Uncertain
