Gene
KWMTBOMO05170
Pre Gene Modal
BGIBMGA012479
Annotation
PREDICTED:_zinc_finger_MIZ_domain-containing_protein_1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.471
Sequence
CDS
ATGCAGAACAACATGACTATGGGGAATCCTCAGTATGGTGCGATGAATGGCTACGGCCAACAGCGCACTCATAATCCGGCCATGACCGGCATGGGAATGGGAGGAAACGGAGGCATGAATGGCATGAACGGTATGAATGGTATGACCGGAATGGGTCAGATGGGCAACGCAGCCATGAATGGTATGAACGGGATGAACCCCATGGCGCAGATGGCTAATATGGGAATGCACGCCAATATGATGCCCGGTCAAATGGGTCCGACACAAATGAGTACTTCAGCAAAAATGGGTCCCAATTACCAAAGGCGACACACGCCATATCCATCTGGAACGATGCTCATGGGACAACGGAAATCCCAGTACATGGGTGGCCAGCCTGGTTTTGGACCGAACCAATACCCCGCCGGATACGGTGGACGGCCCGGCTTCCAAAGTCAATACCCCCCTCAGCAGCCTTTAGGCCCCAGCGGTAATTTCGGAGCTGCAATGAGGGGAACCATAAGACAGTCAACGCCACCTTATACCAATCAAGGACAGTATTTCAATGGCGGAGTTCCCAGTCAATTCCCGCAGCACCAGAGTGCAACAGCGCAGTATGCGGGACAATATAGCGGACAGTTTGCGCAAGAAGTGGCCATGCGAACCAACATGAGTTACCAGCACAGTCCGGTACCGGGTAACCCAACGCCGCCCCTCACCCCCGCCAGCAGTATTCCACCTTACATTAGTCCAAACGGTGACGTAAAACCGCACTTCAACGAGCTGAAACCACCGATGGGCATGGCACAAAACGATGAACTCCGGTTAACATTCCCCGTAAGAGATGGTATTATATTACCGCCATTTAGATTAGAACACAATTTAGCAGTTAGCAACCATGTTTTCCAATTGAAACCCACAGTGCACTCAACGTTGATTTGGAGATCCGACCTTGAGCTGCAGCTAAAATGCTTCCATCATGAAGATCGTCAGATGAACACGAATTGGCCGGCTAGCGTCCAGGTATCGGTGAACGCGACGCCGTTAGTCATAGATAGAGGGGAAAATAAAACATCACACAAGCCATTGTACCTGAAAGAAGTCTGCCAGCCGGGCAGAAACACGATACAGATCACCGTCTCCGCCTGTTGTTGTTCGCATCTATTCGTTCTGCAATTAGTCCACCGGCCGAGCGTACGGAGCGTGTTGCAGGGCTTATTAAGGAAACGGCTGTTGACCGCGGACCATTGCATCGCTAAAATCAAAATGAACTTCAACCAAGCGCCGGCGGTCGGCAACAATGCGAACACTCCAACTGATAGGGACAGCGTCGAACAAACCGCTCTAAAAGTATCATTAAAATGTCCGATCACGTTCAAGAAGATCACACTACCCGCGCGCGGCCACGAGTGCAAACACATACAGTGCTTCGACCTGGAATCTTATCTACAGCTGAATTGCGAACGAGGCTCGTGGAGATGTCCTGTTTGCAACAAACCAGCCCAGCTAGAAGGTCTAGAAGTTGACCAGTACATGTGGGGGATATTGAACACGCTAAACACGTCGGATGTCGATGAAGTGACCATCGACAGCGGAGCTAATTGGAAAGCAGCTAAGAGCCAAAACACGACCGGCATAAAGCAAGAAGACGACAGCGATAACAGCAGCAAACGAGGCAAAGCAATGTCGCCCGGCTCCATGAATATGCCGACCATGAACAACTGGGACATGAACCAAGCGCTGTCGCCGTACCTGCCGCCCGACATGAACACCATCGCTAGCGGCTCCATGATCTCTTCGTACAACCAAAGCGCACAGAACCGAAACGCCGGCCCGAACAGTCAGAACTACGACTTCGGAATGAACAACGGCCCGGGGAGCAATGAGTTCGCCGGCAACGGACCTCTGTCGCACCTCAACGAAAGCGTTAATTCATTAGATCCCCTAAACGCGATGGAGAAGAGCCTAAACGAACAGATGCCGCACACTCCCCACACCCCGCACACGCCGGGGTCGGCGCACACCCCGGGCGGGGGCGGGGGCGCGCACACGCCCGGCTCCTCGCACACGCCGGGCCCGCCCTCTGTCGGACACCACTCGCTCAACGAAGTCGACATTCCAGCCGACCTCAACTTTGACCCCGCGGCTGTCATCGACGGCGAGGGCACGGATAATCTCAATCTGCTTCCGGAAACGAGTGTGGATCCTATGGAGCTGTTATCATATTTGGACGCGCCGGCACTGGGTGAACTGCTCGCGACGCCGCCGTCTTCGTCGTCATCGGCGGGATCGCAGCCACCGCGAGCCCCTTCATCGGACGACCTCTTGGCGCTGTTCGAGTGA
Protein
MQNNMTMGNPQYGAMNGYGQQRTHNPAMTGMGMGGNGGMNGMNGMNGMTGMGQMGNAAMNGMNGMNPMAQMANMGMHANMMPGQMGPTQMSTSAKMGPNYQRRHTPYPSGTMLMGQRKSQYMGGQPGFGPNQYPAGYGGRPGFQSQYPPQQPLGPSGNFGAAMRGTIRQSTPPYTNQGQYFNGGVPSQFPQHQSATAQYAGQYSGQFAQEVAMRTNMSYQHSPVPGNPTPPLTPASSIPPYISPNGDVKPHFNELKPPMGMAQNDELRLTFPVRDGIILPPFRLEHNLAVSNHVFQLKPTVHSTLIWRSDLELQLKCFHHEDRQMNTNWPASVQVSVNATPLVIDRGENKTSHKPLYLKEVCQPGRNTIQITVSACCCSHLFVLQLVHRPSVRSVLQGLLRKRLLTADHCIAKIKMNFNQAPAVGNNANTPTDRDSVEQTALKVSLKCPITFKKITLPARGHECKHIQCFDLESYLQLNCERGSWRCPVCNKPAQLEGLEVDQYMWGILNTLNTSDVDEVTIDSGANWKAAKSQNTTGIKQEDDSDNSSKRGKAMSPGSMNMPTMNNWDMNQALSPYLPPDMNTIASGSMISSYNQSAQNRNAGPNSQNYDFGMNNGPGSNEFAGNGPLSHLNESVNSLDPLNAMEKSLNEQMPHTPHTPHTPGSAHTPGGGGGAHTPGSSHTPGPPSVGHHSLNEVDIPADLNFDPAAVIDGEGTDNLNLLPETSVDPMELLSYLDAPALGELLATPPSSSSSAGSQPPRAPSSDDLLALFE
Summary
Uniprot
H9JSG8
A0A2A4IXM2
A0A194PDI0
A0A212EYT6
A0A194RM72
A0A0L7LC53
+ More
A0A2H1VW96 A0A3S2P454 A0A1Y1MJP6 A0A1B6M2T1 A0A1B6LU89 A0A1B6E294 A0A067QY48 A0A1B6E2J2 A0A2J7PVH9 A0A2J7PVJ0 V9IIX2 D6W8Y8 A0A154PDH1 A0A0T6BG76 V5I9C2 A0A310S8U6 A0A088A5T6 A0A0L7QPG5 A0A2A3E418 A0A1W4XPN0 E2AXE1 A0A1W4XEJ1 A0A1W4XPM2 A0A1W4XE60 A0A195DIE9 F4WBP7 A0A158NRH0 A0A023EXM7 A0A069DX44 A0A195BE49 A0A195FS22 E0VE83 A0A3L8DG24 A0A151XEU0 A0A224XBB3 A0A0K8T0L8 A0A0K8T1L1 A0A0K8T0M1 A0A0K8T0P6 U4UHP7 A0A0C9RCF3 K7IQN2 A0A2L2YTM8 A0A087US88 A0A2P6KJU0 A0A2P6KRV6 A0A2R5LAF2 A0A293LPQ0 A0A2P8Z293 A0A147BU83 A0A131Y0Z1 A0A1E1XK03 A0A131XHW2 L7M6U7 A0A1J1HY39 A0A1E1X4Z9 A0A131Z8Z7 A0A224Z3Z3 A0A131Z4X0 B0W3U7 A0A0L8HCT1 B7P1D3 A0A1Q3EXH8 U5EUB4 Q16PW9 A0A336MBF7 A0A336MAI4 G1N212 K1QVR5 U3JWP8 A0A1B0CG35 A0A091TU59 H0Z7L7 A0A091MEV9 A0A1S3HCU2 A0A091Q292 A0A3B3UJ21 G3U4E9 A0A3B3XS22 A0A3B5LBQ1
A0A2H1VW96 A0A3S2P454 A0A1Y1MJP6 A0A1B6M2T1 A0A1B6LU89 A0A1B6E294 A0A067QY48 A0A1B6E2J2 A0A2J7PVH9 A0A2J7PVJ0 V9IIX2 D6W8Y8 A0A154PDH1 A0A0T6BG76 V5I9C2 A0A310S8U6 A0A088A5T6 A0A0L7QPG5 A0A2A3E418 A0A1W4XPN0 E2AXE1 A0A1W4XEJ1 A0A1W4XPM2 A0A1W4XE60 A0A195DIE9 F4WBP7 A0A158NRH0 A0A023EXM7 A0A069DX44 A0A195BE49 A0A195FS22 E0VE83 A0A3L8DG24 A0A151XEU0 A0A224XBB3 A0A0K8T0L8 A0A0K8T1L1 A0A0K8T0M1 A0A0K8T0P6 U4UHP7 A0A0C9RCF3 K7IQN2 A0A2L2YTM8 A0A087US88 A0A2P6KJU0 A0A2P6KRV6 A0A2R5LAF2 A0A293LPQ0 A0A2P8Z293 A0A147BU83 A0A131Y0Z1 A0A1E1XK03 A0A131XHW2 L7M6U7 A0A1J1HY39 A0A1E1X4Z9 A0A131Z8Z7 A0A224Z3Z3 A0A131Z4X0 B0W3U7 A0A0L8HCT1 B7P1D3 A0A1Q3EXH8 U5EUB4 Q16PW9 A0A336MBF7 A0A336MAI4 G1N212 K1QVR5 U3JWP8 A0A1B0CG35 A0A091TU59 H0Z7L7 A0A091MEV9 A0A1S3HCU2 A0A091Q292 A0A3B3UJ21 G3U4E9 A0A3B3XS22 A0A3B5LBQ1
Pubmed
EMBL
BABH01027855
NWSH01005136
PCG64409.1
KQ459606
KPI91401.1
AGBW02011440
+ More
OWR46656.1 KQ459984 KPJ18923.1 JTDY01001833 KOB72766.1 ODYU01004518 SOQ44484.1 RSAL01000027 RVE51907.1 GEZM01029784 JAV85931.1 GEBQ01009777 JAT30200.1 GEBQ01012741 JAT27236.1 GEDC01005243 JAS32055.1 KK852827 KDR15401.1 GEDC01005145 JAS32153.1 NEVH01020956 PNF20347.1 PNF20349.1 JR046953 AEY60289.1 KQ971312 EEZ98412.2 KQ434878 KZC09893.1 LJIG01000604 KRT86336.1 GALX01003190 JAB65276.1 KQ764027 OAD54634.1 KQ414815 KOC60527.1 KZ288416 PBC26032.1 GL443548 EFN61921.1 KQ980824 KYN12607.1 GL888066 EGI68325.1 ADTU01024005 GBBI01004727 JAC13985.1 GBGD01000424 JAC88465.1 KQ976511 KYM82472.1 KQ981280 KYN43410.1 DS235088 EEB11689.1 QOIP01000008 RLU19400.1 KQ982244 KYQ58845.1 GFTR01008202 JAW08224.1 GBRD01006753 JAG59068.1 GBRD01006756 GDHC01011445 JAG59065.1 JAQ07184.1 GBRD01006754 GDHC01006120 JAG59067.1 JAQ12509.1 GBRD01006752 GDHC01016377 JAG59069.1 JAQ02252.1 KB632360 ERL93519.1 GBYB01005990 GBYB01005991 GBYB01005992 JAG75757.1 JAG75758.1 JAG75759.1 IAAA01050065 LAA11434.1 KK121323 KFM80227.1 MWRG01010285 PRD26605.1 MWRG01006294 PRD29074.1 GGLE01002333 MBY06459.1 GFWV01004080 MAA28810.1 PYGN01000230 PSN50611.1 GEGO01001519 JAR93885.1 GEFM01003651 JAP72145.1 GFAA01003995 JAT99439.1 GEFH01002786 JAP65795.1 GACK01005264 JAA59770.1 CVRI01000035 CRK92867.1 GFAC01004838 JAT94350.1 GEDV01000738 JAP87819.1 GFPF01013442 MAA24588.1 GEDV01003071 JAP85486.1 DS231833 EDS32244.1 KQ418609 KOF86565.1 ABJB010245783 ABJB010704864 ABJB010709924 ABJB010882977 DS616194 EEC00405.1 GFDL01015033 JAV20012.1 GANO01003958 JAB55913.1 CH477769 EAT36429.1 UFQS01000343 UFQT01000343 SSX03021.1 SSX23388.1 UFQT01000806 SSX27352.1 JH816363 EKC40962.1 AGTO01000143 AJWK01010696 AJWK01010697 AJWK01010698 KK405434 KFQ81919.1 ABQF01025410 ABQF01025411 ABQF01025412 KK825649 KFP72552.1 KK684647 KFQ14060.1
OWR46656.1 KQ459984 KPJ18923.1 JTDY01001833 KOB72766.1 ODYU01004518 SOQ44484.1 RSAL01000027 RVE51907.1 GEZM01029784 JAV85931.1 GEBQ01009777 JAT30200.1 GEBQ01012741 JAT27236.1 GEDC01005243 JAS32055.1 KK852827 KDR15401.1 GEDC01005145 JAS32153.1 NEVH01020956 PNF20347.1 PNF20349.1 JR046953 AEY60289.1 KQ971312 EEZ98412.2 KQ434878 KZC09893.1 LJIG01000604 KRT86336.1 GALX01003190 JAB65276.1 KQ764027 OAD54634.1 KQ414815 KOC60527.1 KZ288416 PBC26032.1 GL443548 EFN61921.1 KQ980824 KYN12607.1 GL888066 EGI68325.1 ADTU01024005 GBBI01004727 JAC13985.1 GBGD01000424 JAC88465.1 KQ976511 KYM82472.1 KQ981280 KYN43410.1 DS235088 EEB11689.1 QOIP01000008 RLU19400.1 KQ982244 KYQ58845.1 GFTR01008202 JAW08224.1 GBRD01006753 JAG59068.1 GBRD01006756 GDHC01011445 JAG59065.1 JAQ07184.1 GBRD01006754 GDHC01006120 JAG59067.1 JAQ12509.1 GBRD01006752 GDHC01016377 JAG59069.1 JAQ02252.1 KB632360 ERL93519.1 GBYB01005990 GBYB01005991 GBYB01005992 JAG75757.1 JAG75758.1 JAG75759.1 IAAA01050065 LAA11434.1 KK121323 KFM80227.1 MWRG01010285 PRD26605.1 MWRG01006294 PRD29074.1 GGLE01002333 MBY06459.1 GFWV01004080 MAA28810.1 PYGN01000230 PSN50611.1 GEGO01001519 JAR93885.1 GEFM01003651 JAP72145.1 GFAA01003995 JAT99439.1 GEFH01002786 JAP65795.1 GACK01005264 JAA59770.1 CVRI01000035 CRK92867.1 GFAC01004838 JAT94350.1 GEDV01000738 JAP87819.1 GFPF01013442 MAA24588.1 GEDV01003071 JAP85486.1 DS231833 EDS32244.1 KQ418609 KOF86565.1 ABJB010245783 ABJB010704864 ABJB010709924 ABJB010882977 DS616194 EEC00405.1 GFDL01015033 JAV20012.1 GANO01003958 JAB55913.1 CH477769 EAT36429.1 UFQS01000343 UFQT01000343 SSX03021.1 SSX23388.1 UFQT01000806 SSX27352.1 JH816363 EKC40962.1 AGTO01000143 AJWK01010696 AJWK01010697 AJWK01010698 KK405434 KFQ81919.1 ABQF01025410 ABQF01025411 ABQF01025412 KK825649 KFP72552.1 KK684647 KFQ14060.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000283053 UP000027135 UP000235965 UP000007266 UP000076502 UP000005203 UP000053825 UP000242457 UP000192223 UP000000311 UP000078492 UP000007755 UP000005205 UP000078540 UP000078541 UP000009046 UP000279307 UP000075809 UP000030742 UP000002358 UP000054359 UP000245037 UP000183832 UP000002320 UP000053454 UP000001555 UP000008820 UP000001645 UP000005408 UP000016665 UP000092461 UP000007754 UP000085678 UP000261500 UP000007646 UP000261480 UP000261380
UP000283053 UP000027135 UP000235965 UP000007266 UP000076502 UP000005203 UP000053825 UP000242457 UP000192223 UP000000311 UP000078492 UP000007755 UP000005205 UP000078540 UP000078541 UP000009046 UP000279307 UP000075809 UP000030742 UP000002358 UP000054359 UP000245037 UP000183832 UP000002320 UP000053454 UP000001555 UP000008820 UP000001645 UP000005408 UP000016665 UP000092461 UP000007754 UP000085678 UP000261500 UP000007646 UP000261480 UP000261380
PRIDE
Gene 3D
ProteinModelPortal
H9JSG8
A0A2A4IXM2
A0A194PDI0
A0A212EYT6
A0A194RM72
A0A0L7LC53
+ More
A0A2H1VW96 A0A3S2P454 A0A1Y1MJP6 A0A1B6M2T1 A0A1B6LU89 A0A1B6E294 A0A067QY48 A0A1B6E2J2 A0A2J7PVH9 A0A2J7PVJ0 V9IIX2 D6W8Y8 A0A154PDH1 A0A0T6BG76 V5I9C2 A0A310S8U6 A0A088A5T6 A0A0L7QPG5 A0A2A3E418 A0A1W4XPN0 E2AXE1 A0A1W4XEJ1 A0A1W4XPM2 A0A1W4XE60 A0A195DIE9 F4WBP7 A0A158NRH0 A0A023EXM7 A0A069DX44 A0A195BE49 A0A195FS22 E0VE83 A0A3L8DG24 A0A151XEU0 A0A224XBB3 A0A0K8T0L8 A0A0K8T1L1 A0A0K8T0M1 A0A0K8T0P6 U4UHP7 A0A0C9RCF3 K7IQN2 A0A2L2YTM8 A0A087US88 A0A2P6KJU0 A0A2P6KRV6 A0A2R5LAF2 A0A293LPQ0 A0A2P8Z293 A0A147BU83 A0A131Y0Z1 A0A1E1XK03 A0A131XHW2 L7M6U7 A0A1J1HY39 A0A1E1X4Z9 A0A131Z8Z7 A0A224Z3Z3 A0A131Z4X0 B0W3U7 A0A0L8HCT1 B7P1D3 A0A1Q3EXH8 U5EUB4 Q16PW9 A0A336MBF7 A0A336MAI4 G1N212 K1QVR5 U3JWP8 A0A1B0CG35 A0A091TU59 H0Z7L7 A0A091MEV9 A0A1S3HCU2 A0A091Q292 A0A3B3UJ21 G3U4E9 A0A3B3XS22 A0A3B5LBQ1
A0A2H1VW96 A0A3S2P454 A0A1Y1MJP6 A0A1B6M2T1 A0A1B6LU89 A0A1B6E294 A0A067QY48 A0A1B6E2J2 A0A2J7PVH9 A0A2J7PVJ0 V9IIX2 D6W8Y8 A0A154PDH1 A0A0T6BG76 V5I9C2 A0A310S8U6 A0A088A5T6 A0A0L7QPG5 A0A2A3E418 A0A1W4XPN0 E2AXE1 A0A1W4XEJ1 A0A1W4XPM2 A0A1W4XE60 A0A195DIE9 F4WBP7 A0A158NRH0 A0A023EXM7 A0A069DX44 A0A195BE49 A0A195FS22 E0VE83 A0A3L8DG24 A0A151XEU0 A0A224XBB3 A0A0K8T0L8 A0A0K8T1L1 A0A0K8T0M1 A0A0K8T0P6 U4UHP7 A0A0C9RCF3 K7IQN2 A0A2L2YTM8 A0A087US88 A0A2P6KJU0 A0A2P6KRV6 A0A2R5LAF2 A0A293LPQ0 A0A2P8Z293 A0A147BU83 A0A131Y0Z1 A0A1E1XK03 A0A131XHW2 L7M6U7 A0A1J1HY39 A0A1E1X4Z9 A0A131Z8Z7 A0A224Z3Z3 A0A131Z4X0 B0W3U7 A0A0L8HCT1 B7P1D3 A0A1Q3EXH8 U5EUB4 Q16PW9 A0A336MBF7 A0A336MAI4 G1N212 K1QVR5 U3JWP8 A0A1B0CG35 A0A091TU59 H0Z7L7 A0A091MEV9 A0A1S3HCU2 A0A091Q292 A0A3B3UJ21 G3U4E9 A0A3B3XS22 A0A3B5LBQ1
PDB
4FO9
E-value=3.42413e-17,
Score=219
Ontologies
GO
Topology
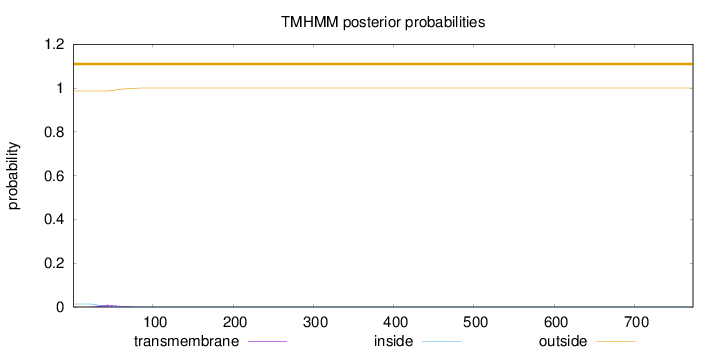
Length:
773
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30027
Exp number, first 60 AAs:
0.24393
Total prob of N-in:
0.01402
outside
1 - 773
Population Genetic Test Statistics
Pi
217.289421
Theta
159.726152
Tajima's D
1.040201
CLR
0
CSRT
0.670816459177041
Interpretation
Uncertain
