Gene
KWMTBOMO05167 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012478
Annotation
PREDICTED:_trypsin_delta/gamma-like_[Bombyx_mori]
Full name
Trypsin-4
+ More
Trypsin-1
Trypsin-2
Trypsin-3
Trypsin 5G1
Trypsin-7
Trypsin-1
Trypsin-2
Trypsin-3
Trypsin 5G1
Trypsin-7
Alternative Name
Antryp1
Antryp2
Antryp2
Location in the cell
Extracellular Reliability : 2.73
Sequence
CDS
ATGAAGTCAAATATTTGTGCCGCTTTTGTTTTTTTCACTTTCTCGACTACTGCTTCAGCATTGCCAACATCTGCAACTTCAAGTGATCTACCATCAAGAATAGTGAATGGTTTTCCGATTGATATAACTGAAATTCCGTACCAAGCTTCCTTGAGACGGAGCGTGGCGTCAGGATGGGCACATTCCTGTGGAGCTGTGATCATTAGCAACAATGCGGTATTGACGGCTGCACACTGTGTTGATTCCTATGTCTTAAATCCTTCAGTTTTAAAAGTAGTCGTTGGAACATCGTACCGACTTTCAGGAGGACAATCCTATGCGATATCGAGAATCATTCCTCACGAACAGTATTTACAATCTACTTTAGAAAATGACATTGCTGTTGTAGGAACATCTAAAACAATGACTTTCAGTGCAAGTGTTAATTCCGTTGCTATTGCTTCATCAACTTTATCTTTACCGATCGGATCTGAAGCACTTGTATCTGGATATGGGACAACATCATACGAAGGCACCACATCATCTGTCCTTCAAGCTGCTAAAGTCAAAGTTATCGCTCAAGAAACGTGTGCTCGGGCGTATTTGCGGATTACCCCGATTACCTCTGGAATGTTATGTGCCAACGCTACCCAGCCTCCAAGGGATGCCTGCCAAGGGGACTCGGGTGGTCCTTTAGTAGCAGACAACGTTCTAATCGGTGTCGTGTCCTGGGGTGAAGGCTGCGCTGATGCTTTATACCCAGGAGTCTACACAAGGGTTTCTGAATATAATTCATGGATTGTGCGCAATGTTGGACTTTTATAA
Protein
MKSNICAAFVFFTFSTTASALPTSATSSDLPSRIVNGFPIDITEIPYQASLRRSVASGWAHSCGAVIISNNAVLTAAHCVDSYVLNPSVLKVVVGTSYRLSGGQSYAISRIIPHEQYLQSTLENDIAVVGTSKTMTFSASVNSVAIASSTLSLPIGSEALVSGYGTTSYEGTTSSVLQAAKVKVIAQETCARAYLRITPITSGMLCANATQPPRDACQGDSGGPLVADNVLIGVVSWGEGCADALYPGVYTRVSEYNSWIVRNVGLL
Summary
Description
Constitutive trypsin that is expressed 2 days after emergence, coinciding with host seeking behavior of the female.
Major function may be to aid in digestion of the blood meal.
Major function may be to aid in digestion of the blood meal.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Digestion
Disulfide bond
Hydrolase
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Trypsin-4
chain Trypsin-4
Uniprot
H9JSG7
A0A194PFC6
A0A194RN29
A0A2H1VE79
A0A0M3T9F2
G9F9I9
+ More
A0A3S2TPW3 A0A212EYY4 A0A0L7L3B0 O16133 A0A182T724 A0A1Y9J0S1 A0A1Y9IS50 A0A2C9GTS6 A0A1Y9GLA7 A0A1Y9HES5 P35038 P35035 A0A240PM69 A0A1Y9J180 A0A1Y9GKV0 A0A1Y9J0J4 P35036 A0A182VGJ0 A0A182U9U3 A0A2C9GTQ1 U5EZ49 A0A2C9GTY9 A0A182G8J0 A0A182MJH4 A0A182TKI7 A0A182M8Z6 U5EZJ4 A0A1Y9GKU7 Q16TL8 A0A182VG56 A0A2K7P6D6 A0A240PL34 A0A182KHH7 C8CGF6 A0A182Y3F4 Q16ID8 A0A240PL37 A0A182T760 Q16TM1 Q0IF85 B8XY09 A0A1Y9J184 A0A1Y9GLX5 A0A182TJQ8 P35037 A0A1S4GXU9 A0A084VY04 E1ZYQ4 A0A084VY03 A0A1B0CUP1 B5AKB2 P29787 A0A240PKC7 A0A182F4C2 A0A1L8DQH5 A0A1L8DQU1 B4LJ08 C5IBF3 A0A084VY00 A4UCN4 A0A1A9XJL9 T1DN08 A0A084VY02 A0A1A9WKF8 A0A182V8D5 A0A1L8DQE6 U5EGR6 A0A1L8DQQ8 A0A1B0BCW7 A0A182F5Y2 A0A1I8PQ52 Q17086 A0A1Q3FP18 G9B5H8 B0WDC9 A0A182TVG7 B0WBW0 A0A1A9VW11 G9B5H7 A0A182GET6 G9B5I4 A0A182GAC9 G9B5J3 G9B5H5 A0A2C9GTS0 B0WF77 G9B5I2 Q16ID4 A0A182F2A5 A0A182L0V6 A0A182XL71 T1DEY9 G9B5H9 P35041
A0A3S2TPW3 A0A212EYY4 A0A0L7L3B0 O16133 A0A182T724 A0A1Y9J0S1 A0A1Y9IS50 A0A2C9GTS6 A0A1Y9GLA7 A0A1Y9HES5 P35038 P35035 A0A240PM69 A0A1Y9J180 A0A1Y9GKV0 A0A1Y9J0J4 P35036 A0A182VGJ0 A0A182U9U3 A0A2C9GTQ1 U5EZ49 A0A2C9GTY9 A0A182G8J0 A0A182MJH4 A0A182TKI7 A0A182M8Z6 U5EZJ4 A0A1Y9GKU7 Q16TL8 A0A182VG56 A0A2K7P6D6 A0A240PL34 A0A182KHH7 C8CGF6 A0A182Y3F4 Q16ID8 A0A240PL37 A0A182T760 Q16TM1 Q0IF85 B8XY09 A0A1Y9J184 A0A1Y9GLX5 A0A182TJQ8 P35037 A0A1S4GXU9 A0A084VY04 E1ZYQ4 A0A084VY03 A0A1B0CUP1 B5AKB2 P29787 A0A240PKC7 A0A182F4C2 A0A1L8DQH5 A0A1L8DQU1 B4LJ08 C5IBF3 A0A084VY00 A4UCN4 A0A1A9XJL9 T1DN08 A0A084VY02 A0A1A9WKF8 A0A182V8D5 A0A1L8DQE6 U5EGR6 A0A1L8DQQ8 A0A1B0BCW7 A0A182F5Y2 A0A1I8PQ52 Q17086 A0A1Q3FP18 G9B5H8 B0WDC9 A0A182TVG7 B0WBW0 A0A1A9VW11 G9B5H7 A0A182GET6 G9B5I4 A0A182GAC9 G9B5J3 G9B5H5 A0A2C9GTS0 B0WF77 G9B5I2 Q16ID4 A0A182F2A5 A0A182L0V6 A0A182XL71 T1DEY9 G9B5H9 P35041
EC Number
3.4.21.4
Pubmed
EMBL
BABH01027869
KQ459606
KPI91399.1
KQ459984
KPJ18922.1
ODYU01002080
+ More
SOQ39135.1 KR024670 ALE15212.1 JN033734 JN315759 AEW46887.1 AFK64823.1 RSAL01000027 RVE51908.1 AGBW02011440 OWR46654.1 JTDY01003207 KOB69947.1 AF012809 AAB66878.1 APCN01000798 Z22930 AAAB01008964 Z18889 Z18890 GANO01001434 JAB58437.1 JXUM01047815 KQ561534 KXJ78237.1 AXCM01007963 AXCM01021675 GANO01000688 JAB59183.1 CH477645 EAT37859.1 GQ398048 ACV07665.1 GQ398049 CH478086 ACV07666.1 EAT34028.1 EAT37856.1 CH477389 EAT42001.1 FJ458410 ACL37992.1 ATLV01018235 KE525226 KFB42848.1 GL435242 EFN73594.1 KFB42847.1 AJWK01029481 EU855138 ACF72874.2 X64363 GFDF01005368 JAV08716.1 GFDF01005369 JAV08715.1 CH940648 EDW60458.2 FJ839343 ACR61324.1 ATLV01018234 KFB42844.1 EF011106 ABM26904.1 GAMD01003270 JAA98320.1 KFB42846.1 GFDF01005398 JAV08686.1 GANO01003334 JAB56537.1 GFDF01005370 JAV08714.1 JXJN01012155 U52359 AAA97479.1 GFDL01005706 JAV29339.1 HM480738 AEI58584.1 DS231896 EDS44442.1 DS231882 EDS42890.1 HM480737 AEI58583.1 JXUM01058527 KQ562007 KXJ76926.1 HM480744 AEI58590.1 JXUM01008849 KQ560270 KXJ83415.1 HM480753 AEI58599.1 HM480735 AEI58581.1 DS231915 EDS26053.1 EDS26054.1 HM480736 HM480742 AEI58588.1 EAT34032.1 GAMD01003326 JAA98264.1 HM480739 AEI58585.1
SOQ39135.1 KR024670 ALE15212.1 JN033734 JN315759 AEW46887.1 AFK64823.1 RSAL01000027 RVE51908.1 AGBW02011440 OWR46654.1 JTDY01003207 KOB69947.1 AF012809 AAB66878.1 APCN01000798 Z22930 AAAB01008964 Z18889 Z18890 GANO01001434 JAB58437.1 JXUM01047815 KQ561534 KXJ78237.1 AXCM01007963 AXCM01021675 GANO01000688 JAB59183.1 CH477645 EAT37859.1 GQ398048 ACV07665.1 GQ398049 CH478086 ACV07666.1 EAT34028.1 EAT37856.1 CH477389 EAT42001.1 FJ458410 ACL37992.1 ATLV01018235 KE525226 KFB42848.1 GL435242 EFN73594.1 KFB42847.1 AJWK01029481 EU855138 ACF72874.2 X64363 GFDF01005368 JAV08716.1 GFDF01005369 JAV08715.1 CH940648 EDW60458.2 FJ839343 ACR61324.1 ATLV01018234 KFB42844.1 EF011106 ABM26904.1 GAMD01003270 JAA98320.1 KFB42846.1 GFDF01005398 JAV08686.1 GANO01003334 JAB56537.1 GFDF01005370 JAV08714.1 JXJN01012155 U52359 AAA97479.1 GFDL01005706 JAV29339.1 HM480738 AEI58584.1 DS231896 EDS44442.1 DS231882 EDS42890.1 HM480737 AEI58583.1 JXUM01058527 KQ562007 KXJ76926.1 HM480744 AEI58590.1 JXUM01008849 KQ560270 KXJ83415.1 HM480753 AEI58599.1 HM480735 AEI58581.1 DS231915 EDS26053.1 EDS26054.1 HM480736 HM480742 AEI58588.1 EAT34032.1 GAMD01003326 JAA98264.1 HM480739 AEI58585.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
+ More
UP000075901 UP000076407 UP000075903 UP000075882 UP000075840 UP000075900 UP000007062 UP000075885 UP000075902 UP000069940 UP000249989 UP000075883 UP000008820 UP000075881 UP000076408 UP000030765 UP000000311 UP000092461 UP000069272 UP000008792 UP000092443 UP000091820 UP000092460 UP000095300 UP000002320 UP000078200
UP000075901 UP000076407 UP000075903 UP000075882 UP000075840 UP000075900 UP000007062 UP000075885 UP000075902 UP000069940 UP000249989 UP000075883 UP000008820 UP000075881 UP000076408 UP000030765 UP000000311 UP000092461 UP000069272 UP000008792 UP000092443 UP000091820 UP000092460 UP000095300 UP000002320 UP000078200
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JSG7
A0A194PFC6
A0A194RN29
A0A2H1VE79
A0A0M3T9F2
G9F9I9
+ More
A0A3S2TPW3 A0A212EYY4 A0A0L7L3B0 O16133 A0A182T724 A0A1Y9J0S1 A0A1Y9IS50 A0A2C9GTS6 A0A1Y9GLA7 A0A1Y9HES5 P35038 P35035 A0A240PM69 A0A1Y9J180 A0A1Y9GKV0 A0A1Y9J0J4 P35036 A0A182VGJ0 A0A182U9U3 A0A2C9GTQ1 U5EZ49 A0A2C9GTY9 A0A182G8J0 A0A182MJH4 A0A182TKI7 A0A182M8Z6 U5EZJ4 A0A1Y9GKU7 Q16TL8 A0A182VG56 A0A2K7P6D6 A0A240PL34 A0A182KHH7 C8CGF6 A0A182Y3F4 Q16ID8 A0A240PL37 A0A182T760 Q16TM1 Q0IF85 B8XY09 A0A1Y9J184 A0A1Y9GLX5 A0A182TJQ8 P35037 A0A1S4GXU9 A0A084VY04 E1ZYQ4 A0A084VY03 A0A1B0CUP1 B5AKB2 P29787 A0A240PKC7 A0A182F4C2 A0A1L8DQH5 A0A1L8DQU1 B4LJ08 C5IBF3 A0A084VY00 A4UCN4 A0A1A9XJL9 T1DN08 A0A084VY02 A0A1A9WKF8 A0A182V8D5 A0A1L8DQE6 U5EGR6 A0A1L8DQQ8 A0A1B0BCW7 A0A182F5Y2 A0A1I8PQ52 Q17086 A0A1Q3FP18 G9B5H8 B0WDC9 A0A182TVG7 B0WBW0 A0A1A9VW11 G9B5H7 A0A182GET6 G9B5I4 A0A182GAC9 G9B5J3 G9B5H5 A0A2C9GTS0 B0WF77 G9B5I2 Q16ID4 A0A182F2A5 A0A182L0V6 A0A182XL71 T1DEY9 G9B5H9 P35041
A0A3S2TPW3 A0A212EYY4 A0A0L7L3B0 O16133 A0A182T724 A0A1Y9J0S1 A0A1Y9IS50 A0A2C9GTS6 A0A1Y9GLA7 A0A1Y9HES5 P35038 P35035 A0A240PM69 A0A1Y9J180 A0A1Y9GKV0 A0A1Y9J0J4 P35036 A0A182VGJ0 A0A182U9U3 A0A2C9GTQ1 U5EZ49 A0A2C9GTY9 A0A182G8J0 A0A182MJH4 A0A182TKI7 A0A182M8Z6 U5EZJ4 A0A1Y9GKU7 Q16TL8 A0A182VG56 A0A2K7P6D6 A0A240PL34 A0A182KHH7 C8CGF6 A0A182Y3F4 Q16ID8 A0A240PL37 A0A182T760 Q16TM1 Q0IF85 B8XY09 A0A1Y9J184 A0A1Y9GLX5 A0A182TJQ8 P35037 A0A1S4GXU9 A0A084VY04 E1ZYQ4 A0A084VY03 A0A1B0CUP1 B5AKB2 P29787 A0A240PKC7 A0A182F4C2 A0A1L8DQH5 A0A1L8DQU1 B4LJ08 C5IBF3 A0A084VY00 A4UCN4 A0A1A9XJL9 T1DN08 A0A084VY02 A0A1A9WKF8 A0A182V8D5 A0A1L8DQE6 U5EGR6 A0A1L8DQQ8 A0A1B0BCW7 A0A182F5Y2 A0A1I8PQ52 Q17086 A0A1Q3FP18 G9B5H8 B0WDC9 A0A182TVG7 B0WBW0 A0A1A9VW11 G9B5H7 A0A182GET6 G9B5I4 A0A182GAC9 G9B5J3 G9B5H5 A0A2C9GTS0 B0WF77 G9B5I2 Q16ID4 A0A182F2A5 A0A182L0V6 A0A182XL71 T1DEY9 G9B5H9 P35041
PDB
1A5I
E-value=6.65216e-29,
Score=315
Ontologies
GO
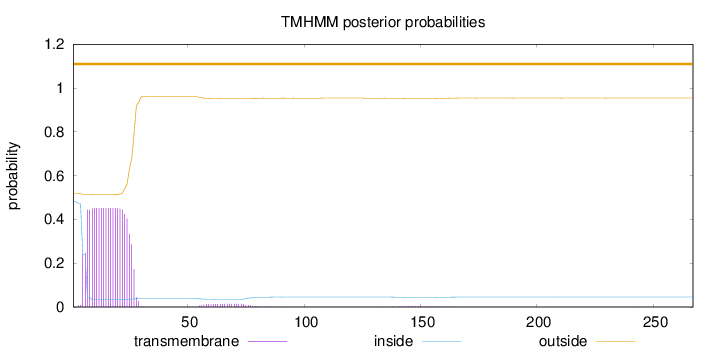
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 20,
Likelihood: 0.982043
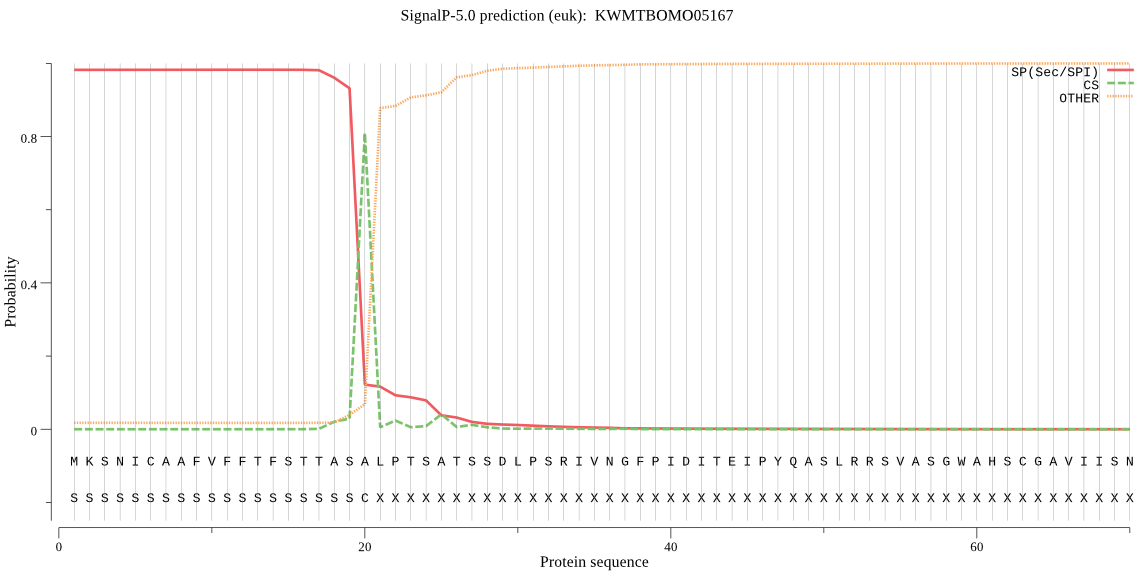
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.81276999999999
Exp number, first 60 AAs:
9.46894
Total prob of N-in:
0.48170
outside
1 - 267
Population Genetic Test Statistics
Pi
201.695388
Theta
144.718641
Tajima's D
0.559807
CLR
0.699122
CSRT
0.533173341332933
Interpretation
Uncertain
