Gene
KWMTBOMO05163
Pre Gene Modal
BGIBMGA012491
Annotation
PREDICTED:_fatty-acid_amide_hydrolase_2-B_isoform_X1_[Amyelois_transitella]
Full name
Cytoplasmic tRNA 2-thiolation protein 2
Location in the cell
Mitochondrial Reliability : 1.937
Sequence
CDS
ATGAGGAGGTTTCTCAGATTTCTCGTATCGTTGCTGGCCATCATAGTGGTGCCGGTTTCATATTTTTCAAGCTTCAAACGAAAACGGAAATGTCCCCCGCCAACAAATCCCATCCTCTTCAAATCAGCAACTAAGCTAGCAGATATGATAAGAAAGAAACAGATAACATCGGAAGAGGTTGTGTCTGCGTTTATAGAACGGTGTAAAGAAGTTAATCCTTACGTAAACGCTGTAATCGAACCACGGTATGAAGTAGCGCTAAGAGAAGCAAGAGGAATAGACAAAATGATAAAGTCAACAGATCGCACGCCAGAAGAATTAGAACAGGAATATCCACTATTGGGAATTCCGCTTACTATCAAAGAAAGTATAGCGGTTGAGGGCATGAGCAATGATTGCGGAACGGTATACACAAATAGAAATCCAGCCAAGAAAGACGCGGCTATTGTCAGGCAAGCCAGAGCGGCTGGTGCAATTCCCATAGCTGTAACCAGTACACCCCAGTTGTGCATGAACTGGGAAACATACAATAATGTTATAGGAGTAACGATGAACCCTTACGATCCGAAACGTACAACTGGAGGCTCTTCGGGGGGTGAGTCGGCTTTGATTTCTTCAGCAGCATCGCTAATAGGACTCGGATCGGATATGGCTGGGTCCCTTAGATTGCCTCCAATGTTCACTGGAATCTATGGACACAAACCATCACCTAGATTACTTTCGATTGAAGGACACATACCAGACTGCTTGGATCCTGATTTTGAAGAATATTTCACTCTTGGTCCTTTAACACGCTACGCCGAAGATTTAGCACTTATGTTAAAAGTTTTAAAGAAACCTGGAAGCCCTGACATACCTTTCGATAAACCGGTTGACGTATCAACACTGAAATTCTATTACATGGAAAGTGATGGCAGTAACGTTTCCGATTCCATTGGTCGTGACGTCAAGATGGCTATGCAAAAAACGCGGGAGTACCTCAAACAAACCTATAATATCGAAGCGAAACCGCTAAGCATAAAAAACATCGAACACATGTGGGAGATAAGCGTCAGAGTATTAATGAATATAAGCCACGTTCGCAACATCTACACTGATCCGAACAATCCTAAGCAATGGGTATCGATCTGGCCTGAAATCATCAAGAAAATGTTTGGTTTATCGAACAACAACTTTTTTTCTGTTTGCTATGGTCCTTTGAAGAAATTCTTTGATGCCCTACCGAGAAGCTACTACAAGAACCTTCTAACTAAATTCGAAGACATTAAAACTGAATTTGAGACAATTTTATCTGACGATGCAGTTTTGTTGTTTCCGACTTTTCCACATCCCGCCCACTTGCACTACCGAGTGTACTATAAATTTTTGAACTGTGGATATTTAACGATGTTCAATGCATTAGGGCTGCCAGTTACGGCCTGTCCGATTGAAATGTCGAAAAAAGGCCTCCCTGTCGGCATCCAAATTGCGGCAAACAGATACAAAGATCATTTAACTGTGGCGGTCGCTAATGAATTTGAGAAAGCATTCGGTGGCTGGGCAGCTCCTAATAAAGATTCGGTTACAGTATAG
Protein
MRRFLRFLVSLLAIIVVPVSYFSSFKRKRKCPPPTNPILFKSATKLADMIRKKQITSEEVVSAFIERCKEVNPYVNAVIEPRYEVALREARGIDKMIKSTDRTPEELEQEYPLLGIPLTIKESIAVEGMSNDCGTVYTNRNPAKKDAAIVRQARAAGAIPIAVTSTPQLCMNWETYNNVIGVTMNPYDPKRTTGGSSGGESALISSAASLIGLGSDMAGSLRLPPMFTGIYGHKPSPRLLSIEGHIPDCLDPDFEEYFTLGPLTRYAEDLALMLKVLKKPGSPDIPFDKPVDVSTLKFYYMESDGSNVSDSIGRDVKMAMQKTREYLKQTYNIEAKPLSIKNIEHMWEISVRVLMNISHVRNIYTDPNNPKQWVSIWPEIIKKMFGLSNNNFFSVCYGPLKKFFDALPRSYYKNLLTKFEDIKTEFETILSDDAVLLFPTFPHPAHLHYRVYYKFLNCGYLTMFNALGLPVTACPIEMSKKGLPVGIQIAANRYKDHLTVAVANEFEKAFGGWAAPNKDSVTV
Summary
Description
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of tRNA(Lys), tRNA(Glu) and tRNA(Gln). May act by forming a heterodimer with NCS6/CTU1 that ligates sulfur from thiocarboxylated URM1 onto the uridine of tRNAs at wobble position.
Similarity
Belongs to the CTU2/NCS2 family.
Feature
chain Cytoplasmic tRNA 2-thiolation protein 2
Uniprot
H9JSI0
A0A2H1V6Y6
A0A2A4J8W8
A0A212F9Z9
A0A194RS76
A0A2A4J9E8
+ More
S4NRR4 A0A0L0CLG6 A0A1B6IHI7 Q7KSA4 Q9I7I6 B4PPP2 A0A0R1E0X8 B3NYX5 B4IIB2 A0A1W4UY27 A0A0P8XYZ4 W8BUC2 B4JXT0 A0A1B6MC63 A0A1I8NV19 A0A0P8ZZU7 A0A0Q9X2J9 B4K7R2 B4M196 A0A0Q9WFY7 A0A0K8W879 A0A034VA70 B4NFW5 A0A0A1XD04 B3M0C2 A0A034VCY2 A0A0A1WEH7 A0A0L7QQF5 A0A0M4ERM4 A0A3B0JQC5 A0A067R9Z7 A0A1B0DF22 A0A1A9UJV6 A0A1B0G122 A0A1B6DM41 B4QS22 A0A1B6CLB9 A0A1B6KZ86 A0A1B6GHK4 A0A1B6GMQ6 A0A1B6GKN2 A0A182Y3R3 A0A0R1E6F6 A0A182PSK5 F4WG47 A0A026WID5 A0A182QF01 A0A1A9W1Q6 A0A158NT29 A0A182NRV4 A0A182RIM1 F5HMI2 A0A182JRZ3 A0A154PFX4 A0A182VZK5 A0A182WSR6 Q7PUX1 A0A2C9GKK9 A0A182IKK7 B4GN21 E2C8F4 A0A182HLL9 Q29BR1 A0A069DUG6 E2AI08 A0A182MS48 B0W356 A0A1B0B703 D6W8X9 A0A1A9YB02 A0A151XBS6 A0A182VKT0 A0A182TGM9 A0A1Q3FF94 Q177C1 A0A224XEL5 A0A1A9Z7F0 A0A1Q3FFE3 A0A195EGG7 T1HYL5 A0A023EUR9 A0A195BG92 A0A195C6L8 A0A182F7I2 A0A023EUW5 A0A2J7PRK9 Q8MZ56 W5JIS9 A0A067R7C9 A0A146KYU1 A0A146LKA9 U5EY65 A0A1W4XEY2
S4NRR4 A0A0L0CLG6 A0A1B6IHI7 Q7KSA4 Q9I7I6 B4PPP2 A0A0R1E0X8 B3NYX5 B4IIB2 A0A1W4UY27 A0A0P8XYZ4 W8BUC2 B4JXT0 A0A1B6MC63 A0A1I8NV19 A0A0P8ZZU7 A0A0Q9X2J9 B4K7R2 B4M196 A0A0Q9WFY7 A0A0K8W879 A0A034VA70 B4NFW5 A0A0A1XD04 B3M0C2 A0A034VCY2 A0A0A1WEH7 A0A0L7QQF5 A0A0M4ERM4 A0A3B0JQC5 A0A067R9Z7 A0A1B0DF22 A0A1A9UJV6 A0A1B0G122 A0A1B6DM41 B4QS22 A0A1B6CLB9 A0A1B6KZ86 A0A1B6GHK4 A0A1B6GMQ6 A0A1B6GKN2 A0A182Y3R3 A0A0R1E6F6 A0A182PSK5 F4WG47 A0A026WID5 A0A182QF01 A0A1A9W1Q6 A0A158NT29 A0A182NRV4 A0A182RIM1 F5HMI2 A0A182JRZ3 A0A154PFX4 A0A182VZK5 A0A182WSR6 Q7PUX1 A0A2C9GKK9 A0A182IKK7 B4GN21 E2C8F4 A0A182HLL9 Q29BR1 A0A069DUG6 E2AI08 A0A182MS48 B0W356 A0A1B0B703 D6W8X9 A0A1A9YB02 A0A151XBS6 A0A182VKT0 A0A182TGM9 A0A1Q3FF94 Q177C1 A0A224XEL5 A0A1A9Z7F0 A0A1Q3FFE3 A0A195EGG7 T1HYL5 A0A023EUR9 A0A195BG92 A0A195C6L8 A0A182F7I2 A0A023EUW5 A0A2J7PRK9 Q8MZ56 W5JIS9 A0A067R7C9 A0A146KYU1 A0A146LKA9 U5EY65 A0A1W4XEY2
Pubmed
19121390
22118469
26354079
23622113
26108605
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 18057021 24495485 25348373 25830018 24845553 25244985 21719571 24508170 21347285 12364791 14747013 17210077 20798317 15632085 26334808 18362917 19820115 17510324 24945155 26483478 20920257 23761445 26823975
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 18057021 24495485 25348373 25830018 24845553 25244985 21719571 24508170 21347285 12364791 14747013 17210077 20798317 15632085 26334808 18362917 19820115 17510324 24945155 26483478 20920257 23761445 26823975
EMBL
BABH01027885
BABH01027886
ODYU01001002
SOQ36610.1
NWSH01002432
PCG68309.1
+ More
AGBW02009539 OWR50561.1 KQ459984 KPJ18921.1 PCG68308.1 GAIX01014342 JAA78218.1 JRES01000232 KNC33140.1 GECU01021339 JAS86367.1 AE014297 AAN14353.1 AAN14354.1 AHN57430.1 BT031317 AAG22162.3 ABY21730.1 CM000160 EDW96142.2 KRK02936.1 CH954181 EDV48378.1 CH480842 EDW49656.1 CH902617 KPU80096.1 KPU80097.1 GAMC01013832 GAMC01013831 GAMC01013830 GAMC01013829 JAB92724.1 CH916377 EDV90492.1 GEBQ01006456 JAT33521.1 KPU80099.1 KPU80100.1 CH933806 KRG02125.1 EDW16433.2 CH940650 EDW67507.1 KRF83322.1 GDHF01021979 GDHF01005100 JAI30335.1 JAI47214.1 GAKP01019538 GAKP01019537 JAC39414.1 CH964251 EDW83182.1 GBXI01005481 JAD08811.1 EDV43128.2 GAKP01019539 JAC39413.1 GBXI01017484 JAC96807.1 KQ414786 KOC60847.1 CP012526 ALC47197.1 OUUW01000007 SPP82582.1 KK852827 KDR15404.1 AJVK01058480 CCAG010008718 GEDC01010553 JAS26745.1 CM000364 EDX12218.1 GEDC01023120 JAS14178.1 GEBQ01023247 JAT16730.1 GECZ01007888 JAS61881.1 GECZ01006054 GECZ01001641 JAS63715.1 JAS68128.1 GECZ01008646 GECZ01006763 JAS61123.1 JAS63006.1 KRK02935.1 GL888128 EGI66814.1 KK107185 EZA55775.1 AXCN02000250 ADTU01025480 ADTU01025481 AAAB01008987 EGK97504.1 KQ434896 KZC10722.1 EAA00887.6 APCN01000909 CH479186 EDW39147.1 GL453666 EFN75789.1 CM000070 EAL26936.5 GBGD01001121 JAC87768.1 GL439621 EFN66916.1 AXCM01000082 DS231830 EDS30764.1 JXJN01009331 KQ971312 EEZ98236.2 KQ982320 KYQ57815.1 GFDL01008820 JAV26225.1 CH477378 EAT42271.1 GFTR01006982 JAW09444.1 GFDL01008823 JAV26222.1 KQ978957 KYN26997.1 ACPB03016841 GAPW01000882 JAC12716.1 KQ976488 KYM83612.1 KQ978205 KYM96482.1 JXUM01115846 JXUM01115847 GAPW01000817 KQ565921 JAC12781.1 KXJ70534.1 NEVH01021956 PNF18975.1 AY113345 AAM29350.1 ADMH02001371 ETN62795.1 KDR15405.1 GDHC01017078 JAQ01551.1 GDHC01010221 JAQ08408.1 GANO01002174 JAB57697.1
AGBW02009539 OWR50561.1 KQ459984 KPJ18921.1 PCG68308.1 GAIX01014342 JAA78218.1 JRES01000232 KNC33140.1 GECU01021339 JAS86367.1 AE014297 AAN14353.1 AAN14354.1 AHN57430.1 BT031317 AAG22162.3 ABY21730.1 CM000160 EDW96142.2 KRK02936.1 CH954181 EDV48378.1 CH480842 EDW49656.1 CH902617 KPU80096.1 KPU80097.1 GAMC01013832 GAMC01013831 GAMC01013830 GAMC01013829 JAB92724.1 CH916377 EDV90492.1 GEBQ01006456 JAT33521.1 KPU80099.1 KPU80100.1 CH933806 KRG02125.1 EDW16433.2 CH940650 EDW67507.1 KRF83322.1 GDHF01021979 GDHF01005100 JAI30335.1 JAI47214.1 GAKP01019538 GAKP01019537 JAC39414.1 CH964251 EDW83182.1 GBXI01005481 JAD08811.1 EDV43128.2 GAKP01019539 JAC39413.1 GBXI01017484 JAC96807.1 KQ414786 KOC60847.1 CP012526 ALC47197.1 OUUW01000007 SPP82582.1 KK852827 KDR15404.1 AJVK01058480 CCAG010008718 GEDC01010553 JAS26745.1 CM000364 EDX12218.1 GEDC01023120 JAS14178.1 GEBQ01023247 JAT16730.1 GECZ01007888 JAS61881.1 GECZ01006054 GECZ01001641 JAS63715.1 JAS68128.1 GECZ01008646 GECZ01006763 JAS61123.1 JAS63006.1 KRK02935.1 GL888128 EGI66814.1 KK107185 EZA55775.1 AXCN02000250 ADTU01025480 ADTU01025481 AAAB01008987 EGK97504.1 KQ434896 KZC10722.1 EAA00887.6 APCN01000909 CH479186 EDW39147.1 GL453666 EFN75789.1 CM000070 EAL26936.5 GBGD01001121 JAC87768.1 GL439621 EFN66916.1 AXCM01000082 DS231830 EDS30764.1 JXJN01009331 KQ971312 EEZ98236.2 KQ982320 KYQ57815.1 GFDL01008820 JAV26225.1 CH477378 EAT42271.1 GFTR01006982 JAW09444.1 GFDL01008823 JAV26222.1 KQ978957 KYN26997.1 ACPB03016841 GAPW01000882 JAC12716.1 KQ976488 KYM83612.1 KQ978205 KYM96482.1 JXUM01115846 JXUM01115847 GAPW01000817 KQ565921 JAC12781.1 KXJ70534.1 NEVH01021956 PNF18975.1 AY113345 AAM29350.1 ADMH02001371 ETN62795.1 KDR15405.1 GDHC01017078 JAQ01551.1 GDHC01010221 JAQ08408.1 GANO01002174 JAB57697.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000037069
UP000000803
+ More
UP000002282 UP000008711 UP000001292 UP000192221 UP000007801 UP000001070 UP000095300 UP000009192 UP000008792 UP000007798 UP000053825 UP000092553 UP000268350 UP000027135 UP000092462 UP000078200 UP000092444 UP000000304 UP000076408 UP000075885 UP000007755 UP000053097 UP000075886 UP000091820 UP000005205 UP000075884 UP000075900 UP000007062 UP000075881 UP000076502 UP000075920 UP000076407 UP000075840 UP000075880 UP000008744 UP000008237 UP000001819 UP000000311 UP000075883 UP000002320 UP000092460 UP000007266 UP000092443 UP000075809 UP000075903 UP000075902 UP000008820 UP000092445 UP000078492 UP000015103 UP000078540 UP000078542 UP000069272 UP000069940 UP000249989 UP000235965 UP000000673 UP000192223
UP000002282 UP000008711 UP000001292 UP000192221 UP000007801 UP000001070 UP000095300 UP000009192 UP000008792 UP000007798 UP000053825 UP000092553 UP000268350 UP000027135 UP000092462 UP000078200 UP000092444 UP000000304 UP000076408 UP000075885 UP000007755 UP000053097 UP000075886 UP000091820 UP000005205 UP000075884 UP000075900 UP000007062 UP000075881 UP000076502 UP000075920 UP000076407 UP000075840 UP000075880 UP000008744 UP000008237 UP000001819 UP000000311 UP000075883 UP000002320 UP000092460 UP000007266 UP000092443 UP000075809 UP000075903 UP000075902 UP000008820 UP000092445 UP000078492 UP000015103 UP000078540 UP000078542 UP000069272 UP000069940 UP000249989 UP000235965 UP000000673 UP000192223
Interpro
SUPFAM
SSF75304
SSF75304
Gene 3D
ProteinModelPortal
H9JSI0
A0A2H1V6Y6
A0A2A4J8W8
A0A212F9Z9
A0A194RS76
A0A2A4J9E8
+ More
S4NRR4 A0A0L0CLG6 A0A1B6IHI7 Q7KSA4 Q9I7I6 B4PPP2 A0A0R1E0X8 B3NYX5 B4IIB2 A0A1W4UY27 A0A0P8XYZ4 W8BUC2 B4JXT0 A0A1B6MC63 A0A1I8NV19 A0A0P8ZZU7 A0A0Q9X2J9 B4K7R2 B4M196 A0A0Q9WFY7 A0A0K8W879 A0A034VA70 B4NFW5 A0A0A1XD04 B3M0C2 A0A034VCY2 A0A0A1WEH7 A0A0L7QQF5 A0A0M4ERM4 A0A3B0JQC5 A0A067R9Z7 A0A1B0DF22 A0A1A9UJV6 A0A1B0G122 A0A1B6DM41 B4QS22 A0A1B6CLB9 A0A1B6KZ86 A0A1B6GHK4 A0A1B6GMQ6 A0A1B6GKN2 A0A182Y3R3 A0A0R1E6F6 A0A182PSK5 F4WG47 A0A026WID5 A0A182QF01 A0A1A9W1Q6 A0A158NT29 A0A182NRV4 A0A182RIM1 F5HMI2 A0A182JRZ3 A0A154PFX4 A0A182VZK5 A0A182WSR6 Q7PUX1 A0A2C9GKK9 A0A182IKK7 B4GN21 E2C8F4 A0A182HLL9 Q29BR1 A0A069DUG6 E2AI08 A0A182MS48 B0W356 A0A1B0B703 D6W8X9 A0A1A9YB02 A0A151XBS6 A0A182VKT0 A0A182TGM9 A0A1Q3FF94 Q177C1 A0A224XEL5 A0A1A9Z7F0 A0A1Q3FFE3 A0A195EGG7 T1HYL5 A0A023EUR9 A0A195BG92 A0A195C6L8 A0A182F7I2 A0A023EUW5 A0A2J7PRK9 Q8MZ56 W5JIS9 A0A067R7C9 A0A146KYU1 A0A146LKA9 U5EY65 A0A1W4XEY2
S4NRR4 A0A0L0CLG6 A0A1B6IHI7 Q7KSA4 Q9I7I6 B4PPP2 A0A0R1E0X8 B3NYX5 B4IIB2 A0A1W4UY27 A0A0P8XYZ4 W8BUC2 B4JXT0 A0A1B6MC63 A0A1I8NV19 A0A0P8ZZU7 A0A0Q9X2J9 B4K7R2 B4M196 A0A0Q9WFY7 A0A0K8W879 A0A034VA70 B4NFW5 A0A0A1XD04 B3M0C2 A0A034VCY2 A0A0A1WEH7 A0A0L7QQF5 A0A0M4ERM4 A0A3B0JQC5 A0A067R9Z7 A0A1B0DF22 A0A1A9UJV6 A0A1B0G122 A0A1B6DM41 B4QS22 A0A1B6CLB9 A0A1B6KZ86 A0A1B6GHK4 A0A1B6GMQ6 A0A1B6GKN2 A0A182Y3R3 A0A0R1E6F6 A0A182PSK5 F4WG47 A0A026WID5 A0A182QF01 A0A1A9W1Q6 A0A158NT29 A0A182NRV4 A0A182RIM1 F5HMI2 A0A182JRZ3 A0A154PFX4 A0A182VZK5 A0A182WSR6 Q7PUX1 A0A2C9GKK9 A0A182IKK7 B4GN21 E2C8F4 A0A182HLL9 Q29BR1 A0A069DUG6 E2AI08 A0A182MS48 B0W356 A0A1B0B703 D6W8X9 A0A1A9YB02 A0A151XBS6 A0A182VKT0 A0A182TGM9 A0A1Q3FF94 Q177C1 A0A224XEL5 A0A1A9Z7F0 A0A1Q3FFE3 A0A195EGG7 T1HYL5 A0A023EUR9 A0A195BG92 A0A195C6L8 A0A182F7I2 A0A023EUW5 A0A2J7PRK9 Q8MZ56 W5JIS9 A0A067R7C9 A0A146KYU1 A0A146LKA9 U5EY65 A0A1W4XEY2
PDB
5H6T
E-value=1.65899e-16,
Score=212
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
SignalP
Position: 1 - 24,
Likelihood: 0.621914
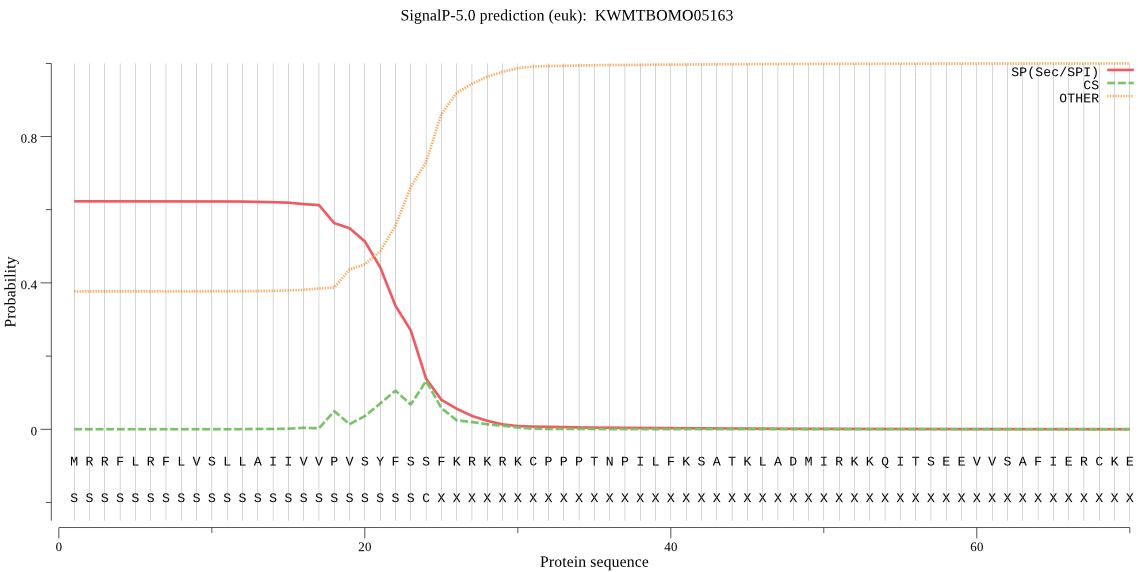
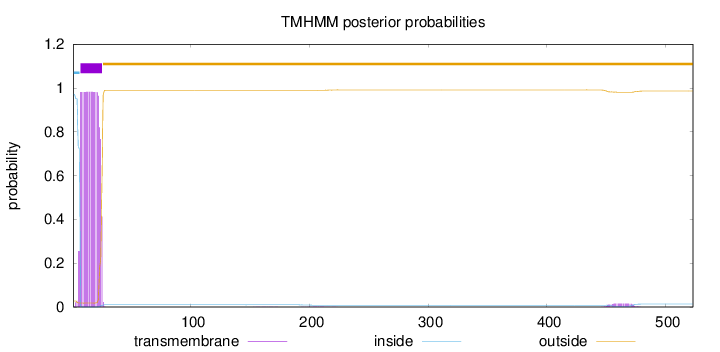
Length:
523
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.66739
Exp number, first 60 AAs:
18.27974
Total prob of N-in:
0.97214
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 25
outside
26 - 523
Population Genetic Test Statistics
Pi
185.828523
Theta
154.838543
Tajima's D
0.755415
CLR
0.628196
CSRT
0.584120793960302
Interpretation
Uncertain
