Gene
KWMTBOMO05160
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.714 PlasmaMembrane Reliability : 1.061
Sequence
CDS
ATGAGTGCAATCAGTTATGTATGTTTTTTACTATTATTAGTAGTTTCCGTTGTTGGTGCTAAAGTTTTGAAAGTAAAATTTGAAGATACGCCGAAATTCGTTGGTTCAGATACAAATGTTACTGTGGTGATCGGAAGAGACGCTACACTTACCTGTACCGTGGAGAACTTGCAAAAATTCAAGGTCGCTTGGCTCAAAGTTGATACACAAACTATCTTGACGATCGGCAATCACGTGATCACGAAGAATCATCGCGTGAGCATCGGCAAAGGAGACCAGACCTGGTCCTTAACGATTCGAGACATTCGTCTGTCAGATGCGGGACAGTACATGTGCCAATTGAACACTGAACCGATGACCATACAGACCCATCATTTACAAGTATCAGTGCCACCGGATATCATAAATTCAAGCAGTAGCGGTGAGATTACAGTACGTGAGGGAGAAAACGTTTCTCTGCATTGCTCTGCTTCTGGTGCTCCCAAACCTACAATCACTTGGAGGAGAGAAGATTTAACTCCAATGAAAATTGGATCTGAAGTGGCGTCGAAATGGAACGGCGTGTGGTTAAACATGTCGTCTGTGAGTCGAGATATTAATGGCGCTTTCCTTTGCATAGCCACAAATGGCGTGCCTCCTTCTGTTAGCAAAAGAATTTTGCTACGTGTTATGTGCAAACCTAAAGTCCATGTAATACAAAAAATGATTGGTGCCTACATCGGAGATACTGTAAATCTTCATTGCAAAATTGAAGCATTTCCACCGCCAGTTATAACCTGGATACATCAAGACACCATCAAATTACAAAATGGTACAAAGTACCAGGTCATCACGGAATCGAAGAGCTACAAACACACAGTGATACTAAGAGTATACAACGTCTGTAGAGAAGACATAGGCTCTTACTATTGCTGCGCTGAGAACCAGCTTGGATCTTCGAAGAACGATGTCACATTATACACCTTAGCAACGACGACTATGAGTACTTCAACAACTGAACGAACCACAACGGCGTGGATCCCGAAATCTACTCAGCCATATGCAGAATTAGAAGTGAGCGTTGATGACGAGGGCCAGAAGTATTCGAATGAAGACAGTTTCGTGGTTGTGCTCAGCCAACAGCAAATGCAAAACATTGGACAAAAGGCAATCATCGATAACTCTAGTTCTATGAAAACAAAATAA
Protein
MSAISYVCFLLLLVVSVVGAKVLKVKFEDTPKFVGSDTNVTVVIGRDATLTCTVENLQKFKVAWLKVDTQTILTIGNHVITKNHRVSIGKGDQTWSLTIRDIRLSDAGQYMCQLNTEPMTIQTHHLQVSVPPDIINSSSSGEITVREGENVSLHCSASGAPKPTITWRREDLTPMKIGSEVASKWNGVWLNMSSVSRDINGAFLCIATNGVPPSVSKRILLRVMCKPKVHVIQKMIGAYIGDTVNLHCKIEAFPPPVITWIHQDTIKLQNGTKYQVITESKSYKHTVILRVYNVCREDIGSYYCCAENQLGSSKNDVTLYTLATTTMSTSTTERTTTAWIPKSTQPYAELEVSVDDEGQKYSNEDSFVVVLSQQQMQNIGQKAIIDNSSSMKTK
Summary
Uniprot
A0A3S2M5M3
A0A2H1V2C2
A0A2A4JGS3
A0A2A4JG82
A0A3S2NEZ3
A0A2H1WX47
+ More
A0A212FD14 A0A0N0P9B5 A0A194PFC1 A0A194QLU6 A0A1A9W2W7 A0A0Q9XDV3 A0A0Q9XGG8 A0A1B0A3T4 A0A0Q9XHX3 A0A0Q9XEG5 B4JC54 A0A0Q9X4T6 A0A3S3QR06 B4KGJ7 B4LRI0 A0A0Q9WIX4 A0A0Q9XHL3 A0A1I8NTJ4 A0A1I8NTJ3 A0A0L0BZ87 B4Q410 A0A3L8D340 A0A0A9YR40 B4MV41 A0A1D2MNL9 Q4V5E0 A0A2H8TXR5 A0A2S2NSZ8 B3MME1 B5DJP7 B4GT22 B3N4Y8 A0A3B0JVB8 A0A146LEP4 A0A2S2Q9R2 A0A0P5I935 A0A0M4E9D1 A0A0J9QW98 A0A0P5EFF7 B4P041 B4I1I8 A0A0N8C4V2 Q9VMN6 A0A3L8E3W8 A0A0A1WQT5 A0A2S2QJB3 A0A2H8TQ35 A0A2S2PKB7 A0A0P5QNC5 A0A0N8DW88 A0A0P5H1B8 A0A1Y1LNF8 A0A1W4VAU1 A0A139WM62 A0A0P5SFZ3 A0A0Q9XG22 A0A0P5MBC8 A0A0P5SYK6 A0A0P6E3M8 A0A0P5ETF1 A0A0Q9XC59 A0A0P5TJ83 B4KGJ5 A0A0P5HVB6 A0A0P5LQL1 A0A0P5J9U8 A0A0M4EBG9 A0A0P6AG16 A0A0C9RKJ2 A0A2H1W6U1 A0A3L8D334 A0A158N9S9 E9FSU5 A0A336KEB9 J9JUC7 A0A139WMQ5 A0A3Q0IW20 A0A0R3NUG6 B4LRH8 B4GT24 D6W8U0 W8BS30 A0A3B0K038 A0A2R5LBW6 A0A154PDG7 A0A1W4VAF1 B4I1I6 E9H6R1 B5DJP9 A0A0R3NUB7 A0A0P5ZTY4
A0A212FD14 A0A0N0P9B5 A0A194PFC1 A0A194QLU6 A0A1A9W2W7 A0A0Q9XDV3 A0A0Q9XGG8 A0A1B0A3T4 A0A0Q9XHX3 A0A0Q9XEG5 B4JC54 A0A0Q9X4T6 A0A3S3QR06 B4KGJ7 B4LRI0 A0A0Q9WIX4 A0A0Q9XHL3 A0A1I8NTJ4 A0A1I8NTJ3 A0A0L0BZ87 B4Q410 A0A3L8D340 A0A0A9YR40 B4MV41 A0A1D2MNL9 Q4V5E0 A0A2H8TXR5 A0A2S2NSZ8 B3MME1 B5DJP7 B4GT22 B3N4Y8 A0A3B0JVB8 A0A146LEP4 A0A2S2Q9R2 A0A0P5I935 A0A0M4E9D1 A0A0J9QW98 A0A0P5EFF7 B4P041 B4I1I8 A0A0N8C4V2 Q9VMN6 A0A3L8E3W8 A0A0A1WQT5 A0A2S2QJB3 A0A2H8TQ35 A0A2S2PKB7 A0A0P5QNC5 A0A0N8DW88 A0A0P5H1B8 A0A1Y1LNF8 A0A1W4VAU1 A0A139WM62 A0A0P5SFZ3 A0A0Q9XG22 A0A0P5MBC8 A0A0P5SYK6 A0A0P6E3M8 A0A0P5ETF1 A0A0Q9XC59 A0A0P5TJ83 B4KGJ5 A0A0P5HVB6 A0A0P5LQL1 A0A0P5J9U8 A0A0M4EBG9 A0A0P6AG16 A0A0C9RKJ2 A0A2H1W6U1 A0A3L8D334 A0A158N9S9 E9FSU5 A0A336KEB9 J9JUC7 A0A139WMQ5 A0A3Q0IW20 A0A0R3NUG6 B4LRH8 B4GT24 D6W8U0 W8BS30 A0A3B0K038 A0A2R5LBW6 A0A154PDG7 A0A1W4VAF1 B4I1I6 E9H6R1 B5DJP9 A0A0R3NUB7 A0A0P5ZTY4
Pubmed
EMBL
RSAL01000027
RVE51914.1
ODYU01000348
SOQ34959.1
NWSH01001540
PCG70918.1
+ More
NWSH01001697 PCG70402.1 RSAL01000150 RVE45783.1 ODYU01011658 SOQ57546.1 AGBW02009133 OWR51588.1 KQ459566 KPI99598.1 KQ459606 KPI91394.1 KQ461198 KPJ05935.1 CH933807 KRG03140.1 KRG03143.1 KRG03139.1 KRG03138.1 CH916368 EDW03067.1 KRG03142.1 NCKU01001248 RWS12630.1 EDW12193.2 CH940649 EDW63575.2 KRF81172.1 KRG03144.1 JRES01001211 KNC24549.1 CM000361 EDX03843.1 QOIP01000014 RLU14912.1 GBHO01011604 JAG32000.1 CH963857 EDW76386.2 LJIJ01000793 ODM94538.1 BT022716 AAY55132.1 GFXV01006153 MBW17958.1 GGMR01007660 MBY20279.1 CH902620 EDV31901.1 KPU73692.1 CH379061 EDY70519.2 KRT04768.1 CH479189 EDW25531.1 CH954177 EDV57890.2 OUUW01000010 SPP86024.1 GDHC01013152 JAQ05477.1 GGMS01005261 MBY74464.1 GDIQ01217857 JAK33868.1 CP012523 ALC38788.1 CM002910 KMY88336.1 KMY88337.1 KMY88338.1 KMY88339.1 KMY88340.1 KMY88341.1 KMY88342.1 GDIQ01270428 GDIQ01270427 JAJ81296.1 CM000157 EDW88906.2 CH480820 EDW54395.1 GDIQ01114643 JAL37083.1 AE014134 BT150446 AAF52276.2 AHZ64220.1 QOIP01000001 RLU27440.1 GBXI01013514 JAD00778.1 GGMS01008621 MBY77824.1 GFXV01004450 MBW16255.1 GGMR01017218 MBY29837.1 GDIQ01269280 GDIQ01132249 JAL19477.1 GDIQ01085764 JAN08973.1 GDIQ01236750 JAK14975.1 GEZM01053974 JAV73890.1 KQ971312 KYB29128.1 GDIP01140200 JAL63514.1 KRG03137.1 GDIQ01166631 JAK85094.1 GDIP01138192 LRGB01003123 JAL65522.1 KZS04428.1 GDIQ01228049 GDIQ01226607 GDIQ01148693 GDIQ01148691 GDIQ01068266 GDIQ01007369 JAN26471.1 GDIQ01270424 GDIQ01270423 JAJ81300.1 KRG03130.1 GDIP01128985 JAL74729.1 EDW12191.2 KRG03129.1 KRG03131.1 KRG03132.1 KRG03133.1 KRG03134.1 KRG03135.1 KRG03136.1 GDIQ01253251 GDIQ01253250 GDIQ01222904 JAK28821.1 GDIQ01166632 JAK85093.1 GDIQ01228048 GDIQ01148692 GDIQ01130937 JAK23677.1 ALC38785.1 GDIP01031348 JAM72367.1 GBYB01013767 JAG83534.1 ODYU01006708 SOQ48801.1 RLU14915.1 ADTU01009347 ADTU01009348 ADTU01009349 ADTU01009350 ADTU01009351 ADTU01009352 ADTU01009353 ADTU01009354 ADTU01009355 ADTU01009356 GL732524 EFX89758.1 UFQS01000363 UFQT01000363 SSX03174.1 SSX23540.1 ABLF02011417 ABLF02011419 ABLF02011420 ABLF02027975 ABLF02027976 ABLF02027977 ABLF02035890 KYB29127.1 KRT04770.1 EDW63573.2 EDW25533.1 EEZ98398.2 GAMC01010479 JAB96076.1 SPP86022.1 GGLE01002819 MBY06945.1 KQ434869 KZC09434.1 EDW54393.1 GL732598 EFX72592.1 EDY70521.2 KRT04769.1 GDIP01042851 JAM60864.1
NWSH01001697 PCG70402.1 RSAL01000150 RVE45783.1 ODYU01011658 SOQ57546.1 AGBW02009133 OWR51588.1 KQ459566 KPI99598.1 KQ459606 KPI91394.1 KQ461198 KPJ05935.1 CH933807 KRG03140.1 KRG03143.1 KRG03139.1 KRG03138.1 CH916368 EDW03067.1 KRG03142.1 NCKU01001248 RWS12630.1 EDW12193.2 CH940649 EDW63575.2 KRF81172.1 KRG03144.1 JRES01001211 KNC24549.1 CM000361 EDX03843.1 QOIP01000014 RLU14912.1 GBHO01011604 JAG32000.1 CH963857 EDW76386.2 LJIJ01000793 ODM94538.1 BT022716 AAY55132.1 GFXV01006153 MBW17958.1 GGMR01007660 MBY20279.1 CH902620 EDV31901.1 KPU73692.1 CH379061 EDY70519.2 KRT04768.1 CH479189 EDW25531.1 CH954177 EDV57890.2 OUUW01000010 SPP86024.1 GDHC01013152 JAQ05477.1 GGMS01005261 MBY74464.1 GDIQ01217857 JAK33868.1 CP012523 ALC38788.1 CM002910 KMY88336.1 KMY88337.1 KMY88338.1 KMY88339.1 KMY88340.1 KMY88341.1 KMY88342.1 GDIQ01270428 GDIQ01270427 JAJ81296.1 CM000157 EDW88906.2 CH480820 EDW54395.1 GDIQ01114643 JAL37083.1 AE014134 BT150446 AAF52276.2 AHZ64220.1 QOIP01000001 RLU27440.1 GBXI01013514 JAD00778.1 GGMS01008621 MBY77824.1 GFXV01004450 MBW16255.1 GGMR01017218 MBY29837.1 GDIQ01269280 GDIQ01132249 JAL19477.1 GDIQ01085764 JAN08973.1 GDIQ01236750 JAK14975.1 GEZM01053974 JAV73890.1 KQ971312 KYB29128.1 GDIP01140200 JAL63514.1 KRG03137.1 GDIQ01166631 JAK85094.1 GDIP01138192 LRGB01003123 JAL65522.1 KZS04428.1 GDIQ01228049 GDIQ01226607 GDIQ01148693 GDIQ01148691 GDIQ01068266 GDIQ01007369 JAN26471.1 GDIQ01270424 GDIQ01270423 JAJ81300.1 KRG03130.1 GDIP01128985 JAL74729.1 EDW12191.2 KRG03129.1 KRG03131.1 KRG03132.1 KRG03133.1 KRG03134.1 KRG03135.1 KRG03136.1 GDIQ01253251 GDIQ01253250 GDIQ01222904 JAK28821.1 GDIQ01166632 JAK85093.1 GDIQ01228048 GDIQ01148692 GDIQ01130937 JAK23677.1 ALC38785.1 GDIP01031348 JAM72367.1 GBYB01013767 JAG83534.1 ODYU01006708 SOQ48801.1 RLU14915.1 ADTU01009347 ADTU01009348 ADTU01009349 ADTU01009350 ADTU01009351 ADTU01009352 ADTU01009353 ADTU01009354 ADTU01009355 ADTU01009356 GL732524 EFX89758.1 UFQS01000363 UFQT01000363 SSX03174.1 SSX23540.1 ABLF02011417 ABLF02011419 ABLF02011420 ABLF02027975 ABLF02027976 ABLF02027977 ABLF02035890 KYB29127.1 KRT04770.1 EDW63573.2 EDW25533.1 EEZ98398.2 GAMC01010479 JAB96076.1 SPP86022.1 GGLE01002819 MBY06945.1 KQ434869 KZC09434.1 EDW54393.1 GL732598 EFX72592.1 EDY70521.2 KRT04769.1 GDIP01042851 JAM60864.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
UP000091820
+ More
UP000009192 UP000092445 UP000001070 UP000285301 UP000008792 UP000095300 UP000037069 UP000000304 UP000279307 UP000007798 UP000094527 UP000007801 UP000001819 UP000008744 UP000008711 UP000268350 UP000092553 UP000002282 UP000001292 UP000000803 UP000192221 UP000007266 UP000076858 UP000005205 UP000000305 UP000007819 UP000079169 UP000076502
UP000009192 UP000092445 UP000001070 UP000285301 UP000008792 UP000095300 UP000037069 UP000000304 UP000279307 UP000007798 UP000094527 UP000007801 UP000001819 UP000008744 UP000008711 UP000268350 UP000092553 UP000002282 UP000001292 UP000000803 UP000192221 UP000007266 UP000076858 UP000005205 UP000000305 UP000007819 UP000079169 UP000076502
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A3S2M5M3
A0A2H1V2C2
A0A2A4JGS3
A0A2A4JG82
A0A3S2NEZ3
A0A2H1WX47
+ More
A0A212FD14 A0A0N0P9B5 A0A194PFC1 A0A194QLU6 A0A1A9W2W7 A0A0Q9XDV3 A0A0Q9XGG8 A0A1B0A3T4 A0A0Q9XHX3 A0A0Q9XEG5 B4JC54 A0A0Q9X4T6 A0A3S3QR06 B4KGJ7 B4LRI0 A0A0Q9WIX4 A0A0Q9XHL3 A0A1I8NTJ4 A0A1I8NTJ3 A0A0L0BZ87 B4Q410 A0A3L8D340 A0A0A9YR40 B4MV41 A0A1D2MNL9 Q4V5E0 A0A2H8TXR5 A0A2S2NSZ8 B3MME1 B5DJP7 B4GT22 B3N4Y8 A0A3B0JVB8 A0A146LEP4 A0A2S2Q9R2 A0A0P5I935 A0A0M4E9D1 A0A0J9QW98 A0A0P5EFF7 B4P041 B4I1I8 A0A0N8C4V2 Q9VMN6 A0A3L8E3W8 A0A0A1WQT5 A0A2S2QJB3 A0A2H8TQ35 A0A2S2PKB7 A0A0P5QNC5 A0A0N8DW88 A0A0P5H1B8 A0A1Y1LNF8 A0A1W4VAU1 A0A139WM62 A0A0P5SFZ3 A0A0Q9XG22 A0A0P5MBC8 A0A0P5SYK6 A0A0P6E3M8 A0A0P5ETF1 A0A0Q9XC59 A0A0P5TJ83 B4KGJ5 A0A0P5HVB6 A0A0P5LQL1 A0A0P5J9U8 A0A0M4EBG9 A0A0P6AG16 A0A0C9RKJ2 A0A2H1W6U1 A0A3L8D334 A0A158N9S9 E9FSU5 A0A336KEB9 J9JUC7 A0A139WMQ5 A0A3Q0IW20 A0A0R3NUG6 B4LRH8 B4GT24 D6W8U0 W8BS30 A0A3B0K038 A0A2R5LBW6 A0A154PDG7 A0A1W4VAF1 B4I1I6 E9H6R1 B5DJP9 A0A0R3NUB7 A0A0P5ZTY4
A0A212FD14 A0A0N0P9B5 A0A194PFC1 A0A194QLU6 A0A1A9W2W7 A0A0Q9XDV3 A0A0Q9XGG8 A0A1B0A3T4 A0A0Q9XHX3 A0A0Q9XEG5 B4JC54 A0A0Q9X4T6 A0A3S3QR06 B4KGJ7 B4LRI0 A0A0Q9WIX4 A0A0Q9XHL3 A0A1I8NTJ4 A0A1I8NTJ3 A0A0L0BZ87 B4Q410 A0A3L8D340 A0A0A9YR40 B4MV41 A0A1D2MNL9 Q4V5E0 A0A2H8TXR5 A0A2S2NSZ8 B3MME1 B5DJP7 B4GT22 B3N4Y8 A0A3B0JVB8 A0A146LEP4 A0A2S2Q9R2 A0A0P5I935 A0A0M4E9D1 A0A0J9QW98 A0A0P5EFF7 B4P041 B4I1I8 A0A0N8C4V2 Q9VMN6 A0A3L8E3W8 A0A0A1WQT5 A0A2S2QJB3 A0A2H8TQ35 A0A2S2PKB7 A0A0P5QNC5 A0A0N8DW88 A0A0P5H1B8 A0A1Y1LNF8 A0A1W4VAU1 A0A139WM62 A0A0P5SFZ3 A0A0Q9XG22 A0A0P5MBC8 A0A0P5SYK6 A0A0P6E3M8 A0A0P5ETF1 A0A0Q9XC59 A0A0P5TJ83 B4KGJ5 A0A0P5HVB6 A0A0P5LQL1 A0A0P5J9U8 A0A0M4EBG9 A0A0P6AG16 A0A0C9RKJ2 A0A2H1W6U1 A0A3L8D334 A0A158N9S9 E9FSU5 A0A336KEB9 J9JUC7 A0A139WMQ5 A0A3Q0IW20 A0A0R3NUG6 B4LRH8 B4GT24 D6W8U0 W8BS30 A0A3B0K038 A0A2R5LBW6 A0A154PDG7 A0A1W4VAF1 B4I1I6 E9H6R1 B5DJP9 A0A0R3NUB7 A0A0P5ZTY4
PDB
6EG1
E-value=4.38973e-72,
Score=690
Ontologies
GO
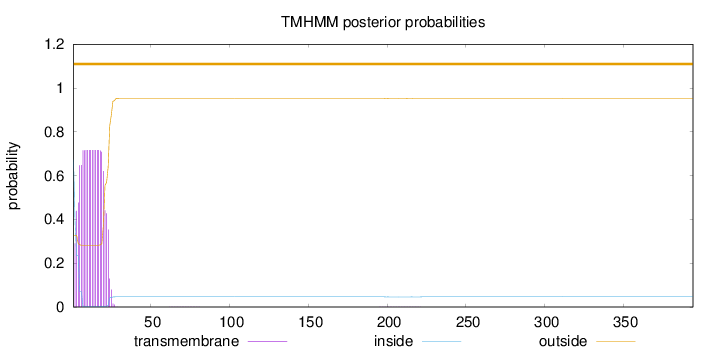
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.978990
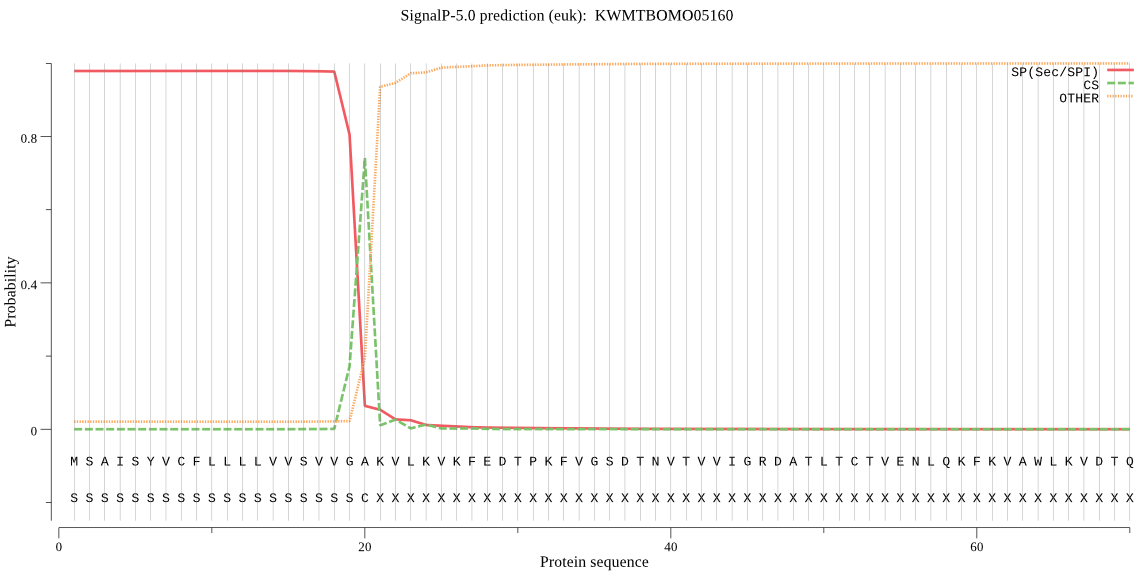
Length:
394
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.91473
Exp number, first 60 AAs:
13.88486
Total prob of N-in:
0.67280
POSSIBLE N-term signal
sequence
outside
1 - 394
Population Genetic Test Statistics
Pi
246.976084
Theta
180.764159
Tajima's D
1.66724
CLR
0.389731
CSRT
0.822508874556272
Interpretation
Uncertain
