Gene
KWMTBOMO05152
Pre Gene Modal
BGIBMGA012495
Annotation
PREDICTED:_speckle-type_POZ_protein_isoform_X1_[Amyelois_transitella]
Full name
Protein roadkill
Alternative Name
Hh-induced MATH and BTB domain-containing protein
Location in the cell
Nuclear Reliability : 1.874
Sequence
CDS
ATGGCCTTAGCCAGGTATAATCAGCAAACCGCGCGCTCCTCTTGGTTGAAGCTCGGTTCGGGCAGCGAGTGCGTTGGCGGGTCGGGCGGCGGCGCGCCCTCTCCGCAGTCGGCGCGCGTCTCTTCGAACGGCGCTGGCGGGATGGCGGTGAGCCGCGTGCCCAGCCCGTTGCACGACGGAAACACGCCCGTCGCAGAAAACTGGTGCTTCACACAAGTCAAAGTGGTGAAATTCAGTTACATGTGGACAATAAACAATTTTAGTTTTTGTAGAGAAGAAATGGGTGAAGTGTTAAAATCATCGACTTTCTCAGCTGGTGCTAGTGATAAATTAAAATGGTGTCTTCGCGTCAATCCAAAAGGACTCGATGAAGAGAGCAAAGACTATTTATCGTTATACTTATTATTAGTCTCGTGTAATAAATCAGAAGTGCGGGCAAAGTTTAAATTTTCAATATTAAATGCTAAAAGAGAAGAAACTAAAGCAATGGAATCGCAACGGGCGTACAGATTCGTGCAAGGAAAAGACTGGGGATTCAAAAAGTTTATCAGAAGAGATTTCCTGCTCGACGAGGCCAATGGTTTATTACCCGAAGATAAACTGACGATATTCTGCGAAGTGTCAGTTGTAGCGGATAGCATCAACATCAGCGGGCAGAGCAACGTGGTTCAGTTCAAAGTGCCTGAGTGCAGATTGTCAGACGATTTAGGAGCTCTCTTTGATAACGAAAGATTCTCTGATGTCACGCTAGCCGTGGGCGGCAGGGAGTTTCAAGCGCATAAAGCTATATTAGCAGCGCGGAGTCCTGTGTTCGCTGCGATGTTCGAACACGAAATGGAAGAGAGGAAGAGGAATAGAGTCGACATAACGGACGTCGATCATGAAGTATTAAGAGAAATGTTACGTTTCATATATACGGGCAGAGCGCCCAATTTGGATAAGATGGCCGACGATCTGCTAGCTGCGGCCGATAAGTATGCATTGGAGCGGCTAAAAGTGATGTGCGAAGAGGCGCTGTGCTTGAGTCTCTCGGTGGAGACTGCCGCAGACACGCTCATACTCGCAGACCTGCACTCCGCCGACCAGCTCAAAGCGCAGACCATAGATTTCATTAACACGAGCCACGCGACGGACGTGATGGACACGGCCGGCTGGAAGAACATGATCTCGTCGCACCCGCACCTCATCGCGGAGGCGTTCCGCGCGCTCGCGACCCAGCAGCAGCAGATCCCGCCCATCGGGCCGCCACGGAAACGTGTGAAGCAGGCCTAG
Protein
MALARYNQQTARSSWLKLGSGSECVGGSGGGAPSPQSARVSSNGAGGMAVSRVPSPLHDGNTPVAENWCFTQVKVVKFSYMWTINNFSFCREEMGEVLKSSTFSAGASDKLKWCLRVNPKGLDEESKDYLSLYLLLVSCNKSEVRAKFKFSILNAKREETKAMESQRAYRFVQGKDWGFKKFIRRDFLLDEANGLLPEDKLTIFCEVSVVADSINISGQSNVVQFKVPECRLSDDLGALFDNERFSDVTLAVGGREFQAHKAILAARSPVFAAMFEHEMEERKRNRVDITDVDHEVLREMLRFIYTGRAPNLDKMADDLLAAADKYALERLKVMCEEALCLSLSVETAADTLILADLHSADQLKAQTIDFINTSHATDVMDTAGWKNMISSHPHLIAEAFRALATQQQQIPPIGPPRKRVKQA
Summary
Description
Involved in segment polarity. In complex with gft/CUL3, promotes ubiquitination of ci and its subsequent degradation by the proteasome, which results in hh signaling attenuation. This regulation may be important during eye formation for proper packing of ommatidia into a hexagonal array.
Subunit
Interacts with ci and gft/CUL3.
Similarity
Belongs to the Tdpoz family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Nucleus
Reference proteome
Segmentation polarity protein
Ubl conjugation pathway
Feature
chain Protein roadkill
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A1E1W5S0
A0A2H1VJN0
A0A212F9Z7
A0A1E1WJF8
A0A194PFB6
A0A1E1VZR2
+ More
A0A194RS66 Q179U2 A0A1J1J0U8 A0A1I8MAR9 A0A1I8Q4G3 A0A2M4CV23 A0A2M4CV51 A0A2M3ZG88 A0A2M4A8E1 T1DCX9 A0A2M3Z1Q4 A0A2M4A8M8 A0A084VI71 A0A1L8DMC3 A0A1L8DM31 A0A1I8JT83 A0A1I8JT76 A0A0K8TMR4 B3LYT1 A0A0A1XH08 A0A336KF12 A0A3B0K1J1 B4NFQ5 Q298W3 A0A2M4A8F9 W8BQW6 F5HK16 A0A1I8MAS0 Q9VFP2-4 Q9VFP2-3 A0A1W4V5V8 A0A1W4USH7 B4R0J2 A0A0R3NH15 I5ANU6 C7LAF6 B4G4S2 A0A0R1E4C9 B4PTD9 A0A0P8YGX7 B4HF69 B3P0K1 Q9VFP2 A0A1W4V4U8 A0A1I8Q4H2 B4MB81 B4JSB8 A0A1A9WJT9 Q7PGC0 B4KC14 A0A0Q9VZH1 A0A139WMR0 A0A0K8V0W6 A0A1Y1NHB6 W8B3B6 A0A0A1WVQ7 A0A1B6DLG6 Q9VFP2-2 A0A1W4US38 I5ANU5 A0A0R1E3Q2 A0A0P8Y1H1 A0A1I8Q4I6 A0A0M3QY84 A0A1B6LAQ1 A0A1B6L6U9 A0A0Q9VZH9 N6U5F7 A0A224XR50 A0A0K8SF32 A0A1B6GH68 A0A023FB09 R4G7P9 A0A0A9Z6U1 A0A0A9Y6H6 A0A0A9Z0Y4 A0A0A9Y1Z4 A0A154PL41 A0A0A9YWX0 V9IDF5 T1JG19 A0A0P6J6Y5 E2C6Z1 A0A151IYV4 A0A0C9RMN6 A0A151WPY5 A0A2A3EEX6 A0A0P5XV92 A0A0C9R9M6 A0A0C9RQU0 A0A158NMX7 A0A224YZW1 A0A131YNK4 A0A3S3PIL6
A0A194RS66 Q179U2 A0A1J1J0U8 A0A1I8MAR9 A0A1I8Q4G3 A0A2M4CV23 A0A2M4CV51 A0A2M3ZG88 A0A2M4A8E1 T1DCX9 A0A2M3Z1Q4 A0A2M4A8M8 A0A084VI71 A0A1L8DMC3 A0A1L8DM31 A0A1I8JT83 A0A1I8JT76 A0A0K8TMR4 B3LYT1 A0A0A1XH08 A0A336KF12 A0A3B0K1J1 B4NFQ5 Q298W3 A0A2M4A8F9 W8BQW6 F5HK16 A0A1I8MAS0 Q9VFP2-4 Q9VFP2-3 A0A1W4V5V8 A0A1W4USH7 B4R0J2 A0A0R3NH15 I5ANU6 C7LAF6 B4G4S2 A0A0R1E4C9 B4PTD9 A0A0P8YGX7 B4HF69 B3P0K1 Q9VFP2 A0A1W4V4U8 A0A1I8Q4H2 B4MB81 B4JSB8 A0A1A9WJT9 Q7PGC0 B4KC14 A0A0Q9VZH1 A0A139WMR0 A0A0K8V0W6 A0A1Y1NHB6 W8B3B6 A0A0A1WVQ7 A0A1B6DLG6 Q9VFP2-2 A0A1W4US38 I5ANU5 A0A0R1E3Q2 A0A0P8Y1H1 A0A1I8Q4I6 A0A0M3QY84 A0A1B6LAQ1 A0A1B6L6U9 A0A0Q9VZH9 N6U5F7 A0A224XR50 A0A0K8SF32 A0A1B6GH68 A0A023FB09 R4G7P9 A0A0A9Z6U1 A0A0A9Y6H6 A0A0A9Z0Y4 A0A0A9Y1Z4 A0A154PL41 A0A0A9YWX0 V9IDF5 T1JG19 A0A0P6J6Y5 E2C6Z1 A0A151IYV4 A0A0C9RMN6 A0A151WPY5 A0A2A3EEX6 A0A0P5XV92 A0A0C9R9M6 A0A0C9RQU0 A0A158NMX7 A0A224YZW1 A0A131YNK4 A0A3S3PIL6
Pubmed
EMBL
GDQN01008738
JAT82316.1
ODYU01002932
SOQ41049.1
AGBW02009539
OWR50554.1
+ More
GDQN01003932 JAT87122.1 KQ459606 KPI91389.1 GDQN01011347 GDQN01010836 JAT79707.1 JAT80218.1 KQ459984 KPJ18911.1 CH477345 EAT43021.1 CVRI01000066 CRL05960.1 GGFL01005014 MBW69192.1 GGFL01004971 MBW69149.1 GGFM01006778 MBW27529.1 GGFK01003752 MBW37073.1 GALA01001652 JAA93200.1 GGFM01001684 MBW22435.1 GGFK01003771 MBW37092.1 ATLV01013303 ATLV01013304 ATLV01013305 ATLV01013306 KE524850 KFB37665.1 GFDF01006567 JAV07517.1 GFDF01006566 JAV07518.1 APCN01002208 GDAI01001961 JAI15642.1 CH902617 EDV44047.1 GBXI01004077 JAD10215.1 UFQS01000350 UFQT01000350 SSX03077.1 SSX23443.1 OUUW01000013 SPP88104.1 CH964251 EDW83122.2 CM000070 EAL27842.2 GGFK01003765 MBW37086.1 GAMC01010934 JAB95621.1 AAAB01008851 EGK96627.1 DQ507278 AE014297 AY121682 BT012443 BT021321 CM000364 EDX13007.1 KRT00308.1 EIM52631.1 BT099706 ACV53070.1 CH479179 EDW24588.1 CM000160 KRK03871.1 EDW97638.1 KPU80594.1 CH480815 EDW42243.1 CH954181 EDV48827.1 CH940656 EDW58352.1 CH916373 EDV94658.1 EAA44989.5 CH933806 EDW16887.2 KRF78249.1 KQ971312 KYB29174.1 GDHF01026604 GDHF01019881 GDHF01017641 GDHF01013322 GDHF01012301 JAI25710.1 JAI32433.1 JAI34673.1 JAI38992.1 JAI40013.1 GEZM01002479 JAV97231.1 GAMC01010935 JAB95620.1 GBXI01013583 GBXI01011687 JAD00709.1 JAD02605.1 GEDC01010791 JAS26507.1 EIM52630.1 KRK03872.1 KPU80595.1 CP012526 ALC47233.1 GEBQ01019231 JAT20746.1 GEBQ01020538 JAT19439.1 KRF78250.1 APGK01048489 KB741112 KB632184 ENN73807.1 ERL89641.1 GFTR01005803 JAW10623.1 GBRD01013929 JAG51897.1 GECZ01007987 JAS61782.1 GBBI01000503 JAC18209.1 ACPB03014368 GAHY01002342 JAA75168.1 GBHO01006079 GBRD01013931 JAG37525.1 JAG51895.1 GBHO01018504 JAG25100.1 GBHO01006078 JAG37526.1 GBHO01018501 GBRD01013930 JAG25103.1 JAG51896.1 KQ434948 KZC12467.1 GBHO01006082 JAG37522.1 JR038652 AEY58359.1 JH432192 GDIQ01022320 JAN72417.1 GL453255 EFN76263.1 KQ980745 KYN13677.1 GBYB01009625 JAG79392.1 KQ982850 KYQ49926.1 KZ288266 PBC30270.1 GDIP01079332 JAM24383.1 GBYB01009627 JAG79394.1 GBYB01009624 GBYB01009626 GBYB01009628 JAG79391.1 JAG79393.1 JAG79395.1 ADTU01020833 ADTU01020834 ADTU01020835 GFPF01012000 MAA23146.1 GEDV01008881 JAP79676.1 NCKU01000330 RWS15969.1
GDQN01003932 JAT87122.1 KQ459606 KPI91389.1 GDQN01011347 GDQN01010836 JAT79707.1 JAT80218.1 KQ459984 KPJ18911.1 CH477345 EAT43021.1 CVRI01000066 CRL05960.1 GGFL01005014 MBW69192.1 GGFL01004971 MBW69149.1 GGFM01006778 MBW27529.1 GGFK01003752 MBW37073.1 GALA01001652 JAA93200.1 GGFM01001684 MBW22435.1 GGFK01003771 MBW37092.1 ATLV01013303 ATLV01013304 ATLV01013305 ATLV01013306 KE524850 KFB37665.1 GFDF01006567 JAV07517.1 GFDF01006566 JAV07518.1 APCN01002208 GDAI01001961 JAI15642.1 CH902617 EDV44047.1 GBXI01004077 JAD10215.1 UFQS01000350 UFQT01000350 SSX03077.1 SSX23443.1 OUUW01000013 SPP88104.1 CH964251 EDW83122.2 CM000070 EAL27842.2 GGFK01003765 MBW37086.1 GAMC01010934 JAB95621.1 AAAB01008851 EGK96627.1 DQ507278 AE014297 AY121682 BT012443 BT021321 CM000364 EDX13007.1 KRT00308.1 EIM52631.1 BT099706 ACV53070.1 CH479179 EDW24588.1 CM000160 KRK03871.1 EDW97638.1 KPU80594.1 CH480815 EDW42243.1 CH954181 EDV48827.1 CH940656 EDW58352.1 CH916373 EDV94658.1 EAA44989.5 CH933806 EDW16887.2 KRF78249.1 KQ971312 KYB29174.1 GDHF01026604 GDHF01019881 GDHF01017641 GDHF01013322 GDHF01012301 JAI25710.1 JAI32433.1 JAI34673.1 JAI38992.1 JAI40013.1 GEZM01002479 JAV97231.1 GAMC01010935 JAB95620.1 GBXI01013583 GBXI01011687 JAD00709.1 JAD02605.1 GEDC01010791 JAS26507.1 EIM52630.1 KRK03872.1 KPU80595.1 CP012526 ALC47233.1 GEBQ01019231 JAT20746.1 GEBQ01020538 JAT19439.1 KRF78250.1 APGK01048489 KB741112 KB632184 ENN73807.1 ERL89641.1 GFTR01005803 JAW10623.1 GBRD01013929 JAG51897.1 GECZ01007987 JAS61782.1 GBBI01000503 JAC18209.1 ACPB03014368 GAHY01002342 JAA75168.1 GBHO01006079 GBRD01013931 JAG37525.1 JAG51895.1 GBHO01018504 JAG25100.1 GBHO01006078 JAG37526.1 GBHO01018501 GBRD01013930 JAG25103.1 JAG51896.1 KQ434948 KZC12467.1 GBHO01006082 JAG37522.1 JR038652 AEY58359.1 JH432192 GDIQ01022320 JAN72417.1 GL453255 EFN76263.1 KQ980745 KYN13677.1 GBYB01009625 JAG79392.1 KQ982850 KYQ49926.1 KZ288266 PBC30270.1 GDIP01079332 JAM24383.1 GBYB01009627 JAG79394.1 GBYB01009624 GBYB01009626 GBYB01009628 JAG79391.1 JAG79393.1 JAG79395.1 ADTU01020833 ADTU01020834 ADTU01020835 GFPF01012000 MAA23146.1 GEDV01008881 JAP79676.1 NCKU01000330 RWS15969.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000008820
UP000183832
UP000095301
+ More
UP000095300 UP000030765 UP000075840 UP000007801 UP000268350 UP000007798 UP000001819 UP000007062 UP000000803 UP000192221 UP000000304 UP000008744 UP000002282 UP000001292 UP000008711 UP000008792 UP000001070 UP000091820 UP000009192 UP000007266 UP000092553 UP000019118 UP000030742 UP000015103 UP000076502 UP000008237 UP000078492 UP000075809 UP000242457 UP000005205 UP000285301
UP000095300 UP000030765 UP000075840 UP000007801 UP000268350 UP000007798 UP000001819 UP000007062 UP000000803 UP000192221 UP000000304 UP000008744 UP000002282 UP000001292 UP000008711 UP000008792 UP000001070 UP000091820 UP000009192 UP000007266 UP000092553 UP000019118 UP000030742 UP000015103 UP000076502 UP000008237 UP000078492 UP000075809 UP000242457 UP000005205 UP000285301
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A1E1W5S0
A0A2H1VJN0
A0A212F9Z7
A0A1E1WJF8
A0A194PFB6
A0A1E1VZR2
+ More
A0A194RS66 Q179U2 A0A1J1J0U8 A0A1I8MAR9 A0A1I8Q4G3 A0A2M4CV23 A0A2M4CV51 A0A2M3ZG88 A0A2M4A8E1 T1DCX9 A0A2M3Z1Q4 A0A2M4A8M8 A0A084VI71 A0A1L8DMC3 A0A1L8DM31 A0A1I8JT83 A0A1I8JT76 A0A0K8TMR4 B3LYT1 A0A0A1XH08 A0A336KF12 A0A3B0K1J1 B4NFQ5 Q298W3 A0A2M4A8F9 W8BQW6 F5HK16 A0A1I8MAS0 Q9VFP2-4 Q9VFP2-3 A0A1W4V5V8 A0A1W4USH7 B4R0J2 A0A0R3NH15 I5ANU6 C7LAF6 B4G4S2 A0A0R1E4C9 B4PTD9 A0A0P8YGX7 B4HF69 B3P0K1 Q9VFP2 A0A1W4V4U8 A0A1I8Q4H2 B4MB81 B4JSB8 A0A1A9WJT9 Q7PGC0 B4KC14 A0A0Q9VZH1 A0A139WMR0 A0A0K8V0W6 A0A1Y1NHB6 W8B3B6 A0A0A1WVQ7 A0A1B6DLG6 Q9VFP2-2 A0A1W4US38 I5ANU5 A0A0R1E3Q2 A0A0P8Y1H1 A0A1I8Q4I6 A0A0M3QY84 A0A1B6LAQ1 A0A1B6L6U9 A0A0Q9VZH9 N6U5F7 A0A224XR50 A0A0K8SF32 A0A1B6GH68 A0A023FB09 R4G7P9 A0A0A9Z6U1 A0A0A9Y6H6 A0A0A9Z0Y4 A0A0A9Y1Z4 A0A154PL41 A0A0A9YWX0 V9IDF5 T1JG19 A0A0P6J6Y5 E2C6Z1 A0A151IYV4 A0A0C9RMN6 A0A151WPY5 A0A2A3EEX6 A0A0P5XV92 A0A0C9R9M6 A0A0C9RQU0 A0A158NMX7 A0A224YZW1 A0A131YNK4 A0A3S3PIL6
A0A194RS66 Q179U2 A0A1J1J0U8 A0A1I8MAR9 A0A1I8Q4G3 A0A2M4CV23 A0A2M4CV51 A0A2M3ZG88 A0A2M4A8E1 T1DCX9 A0A2M3Z1Q4 A0A2M4A8M8 A0A084VI71 A0A1L8DMC3 A0A1L8DM31 A0A1I8JT83 A0A1I8JT76 A0A0K8TMR4 B3LYT1 A0A0A1XH08 A0A336KF12 A0A3B0K1J1 B4NFQ5 Q298W3 A0A2M4A8F9 W8BQW6 F5HK16 A0A1I8MAS0 Q9VFP2-4 Q9VFP2-3 A0A1W4V5V8 A0A1W4USH7 B4R0J2 A0A0R3NH15 I5ANU6 C7LAF6 B4G4S2 A0A0R1E4C9 B4PTD9 A0A0P8YGX7 B4HF69 B3P0K1 Q9VFP2 A0A1W4V4U8 A0A1I8Q4H2 B4MB81 B4JSB8 A0A1A9WJT9 Q7PGC0 B4KC14 A0A0Q9VZH1 A0A139WMR0 A0A0K8V0W6 A0A1Y1NHB6 W8B3B6 A0A0A1WVQ7 A0A1B6DLG6 Q9VFP2-2 A0A1W4US38 I5ANU5 A0A0R1E3Q2 A0A0P8Y1H1 A0A1I8Q4I6 A0A0M3QY84 A0A1B6LAQ1 A0A1B6L6U9 A0A0Q9VZH9 N6U5F7 A0A224XR50 A0A0K8SF32 A0A1B6GH68 A0A023FB09 R4G7P9 A0A0A9Z6U1 A0A0A9Y6H6 A0A0A9Z0Y4 A0A0A9Y1Z4 A0A154PL41 A0A0A9YWX0 V9IDF5 T1JG19 A0A0P6J6Y5 E2C6Z1 A0A151IYV4 A0A0C9RMN6 A0A151WPY5 A0A2A3EEX6 A0A0P5XV92 A0A0C9R9M6 A0A0C9RQU0 A0A158NMX7 A0A224YZW1 A0A131YNK4 A0A3S3PIL6
PDB
3HU6
E-value=2.14913e-130,
Score=1193
Ontologies
GO
GO:0006511
GO:0043161
GO:0030162
GO:0031625
GO:0005634
GO:0005737
GO:0007367
GO:0051865
GO:0042308
GO:0042802
GO:0043065
GO:0042803
GO:0042067
GO:1901044
GO:0005654
GO:0031648
GO:0046330
GO:0045879
GO:0048592
GO:0016567
GO:0001654
GO:0000808
GO:0006260
GO:0005515
GO:0016020
GO:0030170
GO:0007166
GO:0016616
GO:0046168
GO:0016491
GO:0003824
Topology
Subcellular location
Nucleus
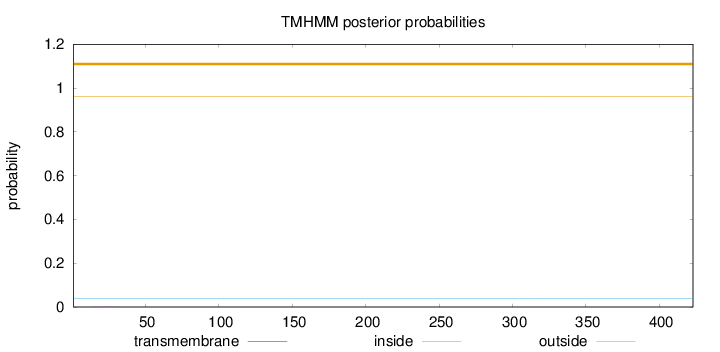
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00614999999999997
Exp number, first 60 AAs:
0.00398
Total prob of N-in:
0.03767
outside
1 - 423
Population Genetic Test Statistics
Pi
218.16571
Theta
183.515188
Tajima's D
0.504474
CLR
0.854007
CSRT
0.512324383780811
Interpretation
Uncertain
