Gene
KWMTBOMO05143
Pre Gene Modal
BGIBMGA012499
Annotation
PREDICTED:_homeobox_protein_ceh-19-like_[Papilio_polytes]
Full name
Homeobox protein ceh-19
Transcription factor
Location in the cell
Nuclear Reliability : 3.781
Sequence
CDS
ATGAATGACAAGAACAACAGAAGACCTCGACGAGCCGGACCTGAACGCAAGCCACGACAAGCTTACAGCGCTAAACAACTAGAGCGATTAGAAGCCGAATTTAAATTGGACAAGTACCTCAGCGTTTCAAAGAGGATGGAACTTTCAAAAGCATTAGGTCTGACAGAAGTACAAATTAAGACCTGGTTCCAGAACCGCAGAACGAAATGGAAGAAGCAGCTAACATCTCGTTTGAAAATCGCACAACGTCAAGGTCTCTTTTCTGGTCAAATCTTCGGACATGCCCCGCAAACTTATTCCCTTTTAAATCCTTACGCGTATGCGCCGCTCAGCTGTGTTTTTACCCCTGTAACATTGCCTTCTACTCCACCATAA
Protein
MNDKNNRRPRRAGPERKPRQAYSAKQLERLEAEFKLDKYLSVSKRMELSKALGLTEVQIKTWFQNRRTKWKKQLTSRLKIAQRQGLFSGQIFGHAPQTYSLLNPYAYAPLSCVFTPVTLPSTPP
Summary
Keywords
Alternative splicing
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Feature
chain Homeobox protein ceh-19
splice variant In isoform a.
splice variant In isoform a.
Uniprot
H9JSI8
A0A2A4IY99
A0A194PEC8
A0A212EN52
A0A2H1V671
A0A0L7KZR4
+ More
A0A1W4WRJ6 D6WAI4 A0A067RRU1 A0A2J7RAG1 A0A2A3E468 N6U3V5 U4UF94 A0A088AEU2 E2C9I3 A0A310S983 A0A1B0CZJ5 A0A0N0BGB2 E9J2H2 A0A151I1J6 F4X620 A0A151WUH1 A0A195C7Q0 A0A158NN00 A0A151IY65 A0A151JZV2 E2ALI4 A0A0C9S2H9 A0A026WY41 A0A0L7R4I2 A0A154PKZ2 A0A232ETK5 A0A336KMZ2 K7IYG2 B0WWU2 A0A0Q9W811 B4K4F7 A0A1J1HRK6 T1HA97 A0A2S2R9R7 J9LIZ8 A0A3L8DKU7 A0A0P8XWM3 A0A0R3P184 P26797-2 A0A261BH12 P26797 A0A2A6CTS8 A0A0N8DXZ1 A0A260ZQI4 A0A2P4W6K9 A0A1I7WMU5 A0A1I8CST3 C3YL06 A0A0B1TRP5
A0A1W4WRJ6 D6WAI4 A0A067RRU1 A0A2J7RAG1 A0A2A3E468 N6U3V5 U4UF94 A0A088AEU2 E2C9I3 A0A310S983 A0A1B0CZJ5 A0A0N0BGB2 E9J2H2 A0A151I1J6 F4X620 A0A151WUH1 A0A195C7Q0 A0A158NN00 A0A151IY65 A0A151JZV2 E2ALI4 A0A0C9S2H9 A0A026WY41 A0A0L7R4I2 A0A154PKZ2 A0A232ETK5 A0A336KMZ2 K7IYG2 B0WWU2 A0A0Q9W811 B4K4F7 A0A1J1HRK6 T1HA97 A0A2S2R9R7 J9LIZ8 A0A3L8DKU7 A0A0P8XWM3 A0A0R3P184 P26797-2 A0A261BH12 P26797 A0A2A6CTS8 A0A0N8DXZ1 A0A260ZQI4 A0A2P4W6K9 A0A1I7WMU5 A0A1I8CST3 C3YL06 A0A0B1TRP5
Pubmed
EMBL
BABH01027930
NWSH01005539
PCG64142.1
KQ459606
KPI91383.1
AGBW02013750
+ More
OWR42881.1 ODYU01000891 SOQ36328.1 JTDY01004152 KOB68524.1 KQ971312 EEZ98636.2 KK852505 KDR22504.1 NEVH01006570 PNF37822.1 KZ288383 PBC26505.1 APGK01043549 KB741015 ENN75301.1 KB632161 ERL89261.1 GL453868 EFN75385.1 KQ763531 OAD55013.1 AJVK01009607 KQ435788 KOX74531.1 GL767845 EFZ12978.1 KQ976560 KYM80588.1 GL888761 EGI58114.1 KQ982729 KYQ51569.1 KQ978231 KYM96173.1 ADTU01020794 KQ980775 KYN13241.1 KQ981374 KYN42388.1 GL440609 EFN65686.1 GBYB01015116 JAG84883.1 KK107072 EZA60636.1 KQ414657 KOC65734.1 KQ434948 KZC12493.1 NNAY01002263 OXU21691.1 UFQS01000487 UFQT01000487 SSX04343.1 SSX24707.1 DS232152 EDS36203.1 CH940657 KRF80925.1 CH933806 EDW13909.2 CVRI01000020 CRK90611.1 ACPB03019798 GGMS01016899 MBY86102.1 ABLF02031382 ABLF02031384 ABLF02031390 QOIP01000007 RLU20792.1 CH902618 KPU79126.1 CH475408 KRT05309.1 Z11794 Z11795 FO080909 NIPN01000140 OZG09493.1 ABKE03000020 PDM81457.1 GDIQ01080940 JAN13797.1 NMWX01000063 OZF87867.1 LFJK02000008 POM45197.1 GG666525 EEN58997.1 KN549303 KHJ98507.1
OWR42881.1 ODYU01000891 SOQ36328.1 JTDY01004152 KOB68524.1 KQ971312 EEZ98636.2 KK852505 KDR22504.1 NEVH01006570 PNF37822.1 KZ288383 PBC26505.1 APGK01043549 KB741015 ENN75301.1 KB632161 ERL89261.1 GL453868 EFN75385.1 KQ763531 OAD55013.1 AJVK01009607 KQ435788 KOX74531.1 GL767845 EFZ12978.1 KQ976560 KYM80588.1 GL888761 EGI58114.1 KQ982729 KYQ51569.1 KQ978231 KYM96173.1 ADTU01020794 KQ980775 KYN13241.1 KQ981374 KYN42388.1 GL440609 EFN65686.1 GBYB01015116 JAG84883.1 KK107072 EZA60636.1 KQ414657 KOC65734.1 KQ434948 KZC12493.1 NNAY01002263 OXU21691.1 UFQS01000487 UFQT01000487 SSX04343.1 SSX24707.1 DS232152 EDS36203.1 CH940657 KRF80925.1 CH933806 EDW13909.2 CVRI01000020 CRK90611.1 ACPB03019798 GGMS01016899 MBY86102.1 ABLF02031382 ABLF02031384 ABLF02031390 QOIP01000007 RLU20792.1 CH902618 KPU79126.1 CH475408 KRT05309.1 Z11794 Z11795 FO080909 NIPN01000140 OZG09493.1 ABKE03000020 PDM81457.1 GDIQ01080940 JAN13797.1 NMWX01000063 OZF87867.1 LFJK02000008 POM45197.1 GG666525 EEN58997.1 KN549303 KHJ98507.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000192223
+ More
UP000007266 UP000027135 UP000235965 UP000242457 UP000019118 UP000030742 UP000005203 UP000008237 UP000092462 UP000053105 UP000078540 UP000007755 UP000075809 UP000078542 UP000005205 UP000078492 UP000078541 UP000000311 UP000053097 UP000053825 UP000076502 UP000215335 UP000002358 UP000002320 UP000008792 UP000009192 UP000183832 UP000015103 UP000007819 UP000279307 UP000007801 UP000001819 UP000001940 UP000216463 UP000232936 UP000216624 UP000237256 UP000095283 UP000095286 UP000001554
UP000007266 UP000027135 UP000235965 UP000242457 UP000019118 UP000030742 UP000005203 UP000008237 UP000092462 UP000053105 UP000078540 UP000007755 UP000075809 UP000078542 UP000005205 UP000078492 UP000078541 UP000000311 UP000053097 UP000053825 UP000076502 UP000215335 UP000002358 UP000002320 UP000008792 UP000009192 UP000183832 UP000015103 UP000007819 UP000279307 UP000007801 UP000001819 UP000001940 UP000216463 UP000232936 UP000216624 UP000237256 UP000095283 UP000095286 UP000001554
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JSI8
A0A2A4IY99
A0A194PEC8
A0A212EN52
A0A2H1V671
A0A0L7KZR4
+ More
A0A1W4WRJ6 D6WAI4 A0A067RRU1 A0A2J7RAG1 A0A2A3E468 N6U3V5 U4UF94 A0A088AEU2 E2C9I3 A0A310S983 A0A1B0CZJ5 A0A0N0BGB2 E9J2H2 A0A151I1J6 F4X620 A0A151WUH1 A0A195C7Q0 A0A158NN00 A0A151IY65 A0A151JZV2 E2ALI4 A0A0C9S2H9 A0A026WY41 A0A0L7R4I2 A0A154PKZ2 A0A232ETK5 A0A336KMZ2 K7IYG2 B0WWU2 A0A0Q9W811 B4K4F7 A0A1J1HRK6 T1HA97 A0A2S2R9R7 J9LIZ8 A0A3L8DKU7 A0A0P8XWM3 A0A0R3P184 P26797-2 A0A261BH12 P26797 A0A2A6CTS8 A0A0N8DXZ1 A0A260ZQI4 A0A2P4W6K9 A0A1I7WMU5 A0A1I8CST3 C3YL06 A0A0B1TRP5
A0A1W4WRJ6 D6WAI4 A0A067RRU1 A0A2J7RAG1 A0A2A3E468 N6U3V5 U4UF94 A0A088AEU2 E2C9I3 A0A310S983 A0A1B0CZJ5 A0A0N0BGB2 E9J2H2 A0A151I1J6 F4X620 A0A151WUH1 A0A195C7Q0 A0A158NN00 A0A151IY65 A0A151JZV2 E2ALI4 A0A0C9S2H9 A0A026WY41 A0A0L7R4I2 A0A154PKZ2 A0A232ETK5 A0A336KMZ2 K7IYG2 B0WWU2 A0A0Q9W811 B4K4F7 A0A1J1HRK6 T1HA97 A0A2S2R9R7 J9LIZ8 A0A3L8DKU7 A0A0P8XWM3 A0A0R3P184 P26797-2 A0A261BH12 P26797 A0A2A6CTS8 A0A0N8DXZ1 A0A260ZQI4 A0A2P4W6K9 A0A1I7WMU5 A0A1I8CST3 C3YL06 A0A0B1TRP5
PDB
2ME6
E-value=1.04428e-10,
Score=153
Ontologies
GO
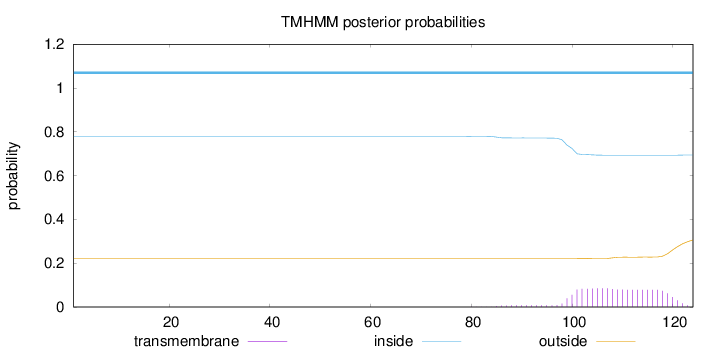
Topology
Subcellular location
Nucleus
Length:
124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.81881
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.77942
inside
1 - 124
Population Genetic Test Statistics
Pi
267.204652
Theta
161.005237
Tajima's D
2.180082
CLR
0
CSRT
0.907954602269887
Interpretation
Uncertain
