Gene
KWMTBOMO05141
Pre Gene Modal
BGIBMGA014340
Annotation
PREDICTED:_striatin-interacting_protein_1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.164 Nuclear Reliability : 2.002
Sequence
CDS
ATGGGTACCAACGGCAATGGTCGTCGCGGAGGTACTAAATTACCGGATGTTCGCCGTCCCCTGGAAGAAGGAGATTGCGTTAACGACATGGAGAGCCCCGAGCTTGACTTCGTCTACGATGACTGTGATACTCAAGCCAACGAAATTGCTGAAATATATAGTTACACGGAACATCCCGAATATCAGCTAAACGTTAAGGCTTTTGAGGATATAATGGAAGAATTTAATCATCCACCGAGTTGGCAAAGACTAACAAAAAAACAACAACAAACTGTAGTAATGATGCTATTGGAACATCTCGATGTAGCTATCTCCGAAATTCGTATGCGTGCTATTCGTTGCATTCTCTATCTGGCCCAAGGGTGCTGGGCAGAAATGCAATCGGATGCCGAACAAGGACATTGGGCTCGAGTGAACATTATGACATTATATGAAATGGGAACATTTTCAATATTTGTGGAATTGCTCAACCTCGAGATTGTTCATAGTAGCTCAGCTATAGCTTCAAGAAAATTGGCAGAATCGTTAGCTGACTCTACAAACTTGCGAGTGATACTGACAGTTCTATATCTTATAACTGAATACATGAGAACAGAGAAAAACAGCCCAGATTCTGAGTATGCACATCTCGTGAAAAATTTTACTGAAGAATTAGGCACACCATTTGGTGATGAGTTGTTACCTGTGAAACTATTAAGTATGGTTACGCATTTGTGTGCTGGGACTACACCACACTTTCCAATGAAGAAAGTTCTCTTACTTTTGTGGAAAGTGTTATTAGTTTACTTGGGTGGCTCAAAGGAACTAAAAGAAAGAAAAGCAAAGTTGAGGGAGCAGCATGGGCTCCCTCCCATCAGTGAAGATACGTTGGAAGTAATAAAGGGAATGAGAGCCAGTTCACCACCTCCAAATGCTGCAGATGTGTTAGAAAATCAAAATCCTAGTCAAAGAAAAGTAAAGGAAAATGAAAGAGCCCTGAGGAGACAAACATTTACTAAACAGACTTCTCTAGATGAAACTGATGAACAAATATTTGTTGACAAAGAAGACACTGGAAATGGATCTGAAGATTATCCTATGGAGTTTCAGTCAATGTCAAGTGACAATCAGCAAGGTGAAAATAGCCAAAATTGTCCATATTATGTATACTTTAAACAGTTGGATAGTCCACCACCCCCACCGCCACTACCCCGTAGTCTGCCATGGCAGTCAAAGGTTCGTCAAAAAGATATTGATATGTTTCTAGATAATGTCCGCATTAAGTTTGTAGGCTATTCTTTACCAGGTGATAGACAAACAATAGCTGGACTGCCACAGCCCATCCATGAGGGTATTGAGATATTAAAGAAACACATGTACACAAGTCTATCAGAAATCCAAGTTGAGAGAGAAATTGAGATTGCATGGAGTCCTCTTACTAGGGGAGAAAAATATGTAGAAGAAACTGAAGCAGAAATTCTATATAGAGCTATGCTACCTAATTTGCCTCAATACATGATTGCTTTGCTAAAGATACTCTTGGCTGCAGCTCCCACTTCAACTGCAAAAACATATTCTATTAATATTATGGCTGATTTGTTACCTGAAGAAATGCCCATGACAGTTCTCCAATCCTTAAAACTAGGCATAGATGTAAACAGACACAAAGAAATTATTGTGAAAGCTGTCACAGCAATCCTATTACTCCTCTTGAAGCATTTTAAACTAAATCATGTCTACCAATTTGAATTTATGTCACAACATTTAGTTTATGCAAACTGTATGCCGCTGGTTCTCAAGTTCTTCAATCAAAACATACTGTCTTATGTAGGAGCCAAGAATTCCATCCCAATATTTGATTTCCCTGCTTGTGTTATTGGTGAACAACCAGAATTAACTAAGGAATATTTAGACATTGGAGATTCCTCTGTTCCATATTCGTGGAGAAATGTATTCTCATGCATAAACTTATTGAGAATTTTGAATAAATTAACAAAATGGAAGAATGCTAGGATCATGATGCTAGTTGTATTCAAAAGCGCACCAATTTTGAAAAGAACATTAAAAGTAAGACATGCCTTGATGCAGTTTTATGTACTGAAATTACTAAAAATGCAAACTAAATATTTAGGAAGACAGTGGCGAAAAACAAACATGAAAACAATAAGCGCAATATATTCAAAAGTTCGTCATCGATTAAATGATGATTGGGCATTTGGAAATGATGTAGATGCAAGACCTTGGGACTTTCAAAGTGAGGAATGTGCATTAAGAATCAGTGTAGACAGATTCAACCACAGGAGATATGGTACAGGTACAGAACAAGAAATTGATCTGACTCCCTTGGACACAGACATAAACAGTGTCCTGGAGAGTAACATAGAGTTAGATGAAGAGTTCAAAGAGAATTATGAATTGTGGCTCGAACAAGAAGTCTATAATAATGAGATCAACTGGGATGTCTTATTATCTACATAG
Protein
MGTNGNGRRGGTKLPDVRRPLEEGDCVNDMESPELDFVYDDCDTQANEIAEIYSYTEHPEYQLNVKAFEDIMEEFNHPPSWQRLTKKQQQTVVMMLLEHLDVAISEIRMRAIRCILYLAQGCWAEMQSDAEQGHWARVNIMTLYEMGTFSIFVELLNLEIVHSSSAIASRKLAESLADSTNLRVILTVLYLITEYMRTEKNSPDSEYAHLVKNFTEELGTPFGDELLPVKLLSMVTHLCAGTTPHFPMKKVLLLLWKVLLVYLGGSKELKERKAKLREQHGLPPISEDTLEVIKGMRASSPPPNAADVLENQNPSQRKVKENERALRRQTFTKQTSLDETDEQIFVDKEDTGNGSEDYPMEFQSMSSDNQQGENSQNCPYYVYFKQLDSPPPPPPLPRSLPWQSKVRQKDIDMFLDNVRIKFVGYSLPGDRQTIAGLPQPIHEGIEILKKHMYTSLSEIQVEREIEIAWSPLTRGEKYVEETEAEILYRAMLPNLPQYMIALLKILLAAAPTSTAKTYSINIMADLLPEEMPMTVLQSLKLGIDVNRHKEIIVKAVTAILLLLLKHFKLNHVYQFEFMSQHLVYANCMPLVLKFFNQNILSYVGAKNSIPIFDFPACVIGEQPELTKEYLDIGDSSVPYSWRNVFSCINLLRILNKLTKWKNARIMMLVVFKSAPILKRTLKVRHALMQFYVLKLLKMQTKYLGRQWRKTNMKTISAIYSKVRHRLNDDWAFGNDVDARPWDFQSEECALRISVDRFNHRRYGTGTEQEIDLTPLDTDINSVLESNIELDEEFKENYELWLEQEVYNNEINWDVLLST
Summary
Uniprot
A0A194RNB1
A0A194PDG3
A0A2A4IY94
A0A212EN26
A0A3S2PHY4
A0A2H1V7H1
+ More
A0A0L7KP37 D6W7U6 A0A1W4X2N7 A0A1W4XDG2 A0A1Y1MPC4 K7IZ64 Q170P2 A0A1S4FHS4 A0A1S4J3C9 E9IPU7 A0A1S4J3S8 A0A0C9QNU7 A0A026VWI0 A0A0J7KQ46 A0A151IW48 A0A195F4X1 A0A151WNC6 E1ZY19 A0A195BEJ5 A0A158NQU1 F4WB84 U5EY75 A0A195C972 A0A1B6KHM4 A0A1B6GDB8 A0A1B6GVD4 A0A1B6JR58 A0A067R907 T1I949 A0A2P8ZK48 A0A182IZI2 A0A023F3F3 A0A224XF16 A0A0N0U536 A0A088A5L4 A0A2A3EU62 A0A2R7X1W1 A0A0A9W8Z8 A0A154P6P5 A0A336LIW2 A0A182PMD6 A0A1J1I572 B0W511 A0A310SDB1 E0VIM0 A0A1S4GXD8 Q7Q3J1 A0A182HVA7 A0A0N8BQQ6 A0A0P6G800 E2BDC5 A0A0N8E5Y4 A0A0P6EIG3 A0A0P6AQL9 E9GEM8 A0A0N8CZ07 A0A0P5MZI4 A0A0N8BB95 A0A0P5XII2 A0A0P5V3K0 A0A0P4YXY2 A0A0P5CR49 A0A0P5D5Z6 A0A0P5BTN9 A0A0P5U0P3 A0A0P4YQA0 A0A0P5U1L2 A0A0P6DA04 A0A0P4XQF4 A0A0N8CL03 A0A0L7QRX1 A0A336LN51 A0A1A9UJF2 A0A1A9W390 A0A1A9Z2P8 A0A1A9YEN6 A0A1B0B192 A0A0R3P5T1 T1JAT6 A0A0R3P914 A0A1I8Q8A8 T1P919 A0A0M3QW90 A0A0L0CHY1 Q29ET1 B4L983 B4LBT0 K1QH32 B4N556 B3M4I5 A0A0N8C4V4 Q8IRD5
A0A0L7KP37 D6W7U6 A0A1W4X2N7 A0A1W4XDG2 A0A1Y1MPC4 K7IZ64 Q170P2 A0A1S4FHS4 A0A1S4J3C9 E9IPU7 A0A1S4J3S8 A0A0C9QNU7 A0A026VWI0 A0A0J7KQ46 A0A151IW48 A0A195F4X1 A0A151WNC6 E1ZY19 A0A195BEJ5 A0A158NQU1 F4WB84 U5EY75 A0A195C972 A0A1B6KHM4 A0A1B6GDB8 A0A1B6GVD4 A0A1B6JR58 A0A067R907 T1I949 A0A2P8ZK48 A0A182IZI2 A0A023F3F3 A0A224XF16 A0A0N0U536 A0A088A5L4 A0A2A3EU62 A0A2R7X1W1 A0A0A9W8Z8 A0A154P6P5 A0A336LIW2 A0A182PMD6 A0A1J1I572 B0W511 A0A310SDB1 E0VIM0 A0A1S4GXD8 Q7Q3J1 A0A182HVA7 A0A0N8BQQ6 A0A0P6G800 E2BDC5 A0A0N8E5Y4 A0A0P6EIG3 A0A0P6AQL9 E9GEM8 A0A0N8CZ07 A0A0P5MZI4 A0A0N8BB95 A0A0P5XII2 A0A0P5V3K0 A0A0P4YXY2 A0A0P5CR49 A0A0P5D5Z6 A0A0P5BTN9 A0A0P5U0P3 A0A0P4YQA0 A0A0P5U1L2 A0A0P6DA04 A0A0P4XQF4 A0A0N8CL03 A0A0L7QRX1 A0A336LN51 A0A1A9UJF2 A0A1A9W390 A0A1A9Z2P8 A0A1A9YEN6 A0A1B0B192 A0A0R3P5T1 T1JAT6 A0A0R3P914 A0A1I8Q8A8 T1P919 A0A0M3QW90 A0A0L0CHY1 Q29ET1 B4L983 B4LBT0 K1QH32 B4N556 B3M4I5 A0A0N8C4V4 Q8IRD5
Pubmed
26354079
22118469
26227816
18362917
19820115
28004739
+ More
20075255 17510324 21282665 24508170 30249741 20798317 21347285 21719571 24845553 29403074 25474469 25401762 26823975 20566863 12364791 21292972 15632085 17994087 23185243 25315136 26108605 22992520 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
20075255 17510324 21282665 24508170 30249741 20798317 21347285 21719571 24845553 29403074 25474469 25401762 26823975 20566863 12364791 21292972 15632085 17994087 23185243 25315136 26108605 22992520 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
KQ459984
KPJ18904.1
KQ459606
KPI91381.1
NWSH01005539
PCG64140.1
+ More
AGBW02013750 OWR42879.1 RSAL01000027 RVE51926.1 ODYU01000891 SOQ36332.1 JTDY01007635 KOB65057.1 KQ971307 EFA11227.2 GEZM01028000 JAV86530.1 CH477471 EAT40408.1 GL764626 EFZ17426.1 GBYB01002284 GBYB01002287 GBYB01002289 GBYB01008691 JAG72051.1 JAG72054.1 JAG72056.1 JAG78458.1 KK107678 QOIP01000004 EZA48128.1 RLU24195.1 LBMM01004386 KMQ92477.1 KQ980874 KYN12049.1 KQ981810 KYN35441.1 KQ982907 KYQ49379.1 GL435171 EFN73868.1 KQ976500 KYM83011.1 ADTU01023603 GL888059 EGI68535.1 GANO01000473 JAB59398.1 KQ978068 KYM97414.1 GEBQ01029009 JAT10968.1 GECZ01019726 GECZ01009362 JAS50043.1 JAS60407.1 GECZ01003388 JAS66381.1 GECU01005987 JAT01720.1 KK852855 KDR14931.1 ACPB03021415 PYGN01000032 PSN56869.1 GBBI01002869 JAC15843.1 GFTR01008048 JAW08378.1 KQ435794 KOX73808.1 KZ288189 PBC34591.1 KK856447 PTY25774.1 GBHO01042284 GDHC01019872 JAG01320.1 JAP98756.1 KQ434827 KZC07616.1 UFQT01000016 SSX17796.1 CVRI01000042 CRK95487.1 DS231840 EDS34577.1 KQ767803 OAD53293.1 DS235201 EEB13226.1 AAAB01008964 EAA12917.4 APCN01002503 GDIQ01154211 JAK97514.1 GDIQ01055902 JAN38835.1 GL447585 EFN86353.1 GDIQ01058596 JAN36141.1 GDIQ01076035 JAN18702.1 GDIP01039643 JAM64072.1 GL732541 EFX82053.1 GDIP01235567 GDIP01229618 GDIP01196221 GDIP01195069 GDIP01191769 GDIP01185981 GDIP01181826 GDIP01181825 GDIP01180075 GDIP01180074 GDIP01154585 GDIP01146329 GDIQ01239492 GDIP01130987 GDIP01130986 GDIP01130985 GDIP01114239 GDIP01108129 GDIP01095893 GDIP01095892 GDIP01094690 GDIP01094689 GDIP01094688 GDIP01084991 GDIP01083245 GDIP01074392 GDIP01073145 GDIP01073144 GDIP01073095 GDIP01071684 GDIP01071683 GDIP01070142 GDIP01062993 GDIP01062992 GDIP01062991 GDIP01051047 GDIP01051046 GDIP01044050 GDIP01044049 GDIP01044048 GDIQ01081938 GDIQ01081937 GDIQ01035017 LRGB01002124 JAK12233.1 JAM18724.1 KZS08999.1 GDIQ01151311 JAL00415.1 GDIQ01194699 JAK57026.1 GDIP01229619 GDIP01195068 GDIP01195067 GDIP01191770 GDIP01185982 GDIP01181828 GDIP01180077 GDIP01177742 GDIP01146330 GDIP01130988 GDIP01114285 GDIP01114240 GDIP01110183 GDIP01108132 GDIP01105683 GDIP01095891 GDIP01094692 GDIP01084990 GDIP01083244 GDIP01073094 GDIP01071682 GDIP01070141 GDIP01062990 GDIP01044047 JAM32033.1 GDIP01105684 JAL98030.1 GDIP01225467 JAI97934.1 GDIP01167966 JAJ55436.1 GDIP01239540 GDIP01238228 GDIP01238227 GDIP01236780 GDIP01231025 GDIP01231024 GDIP01231023 GDIP01227817 GDIP01220980 GDIP01220979 GDIP01210470 GDIP01201877 GDIP01201876 GDIP01201875 GDIP01200619 GDIP01200618 GDIP01200617 GDIP01197278 GDIP01197277 GDIP01197274 GDIP01167023 GDIP01160373 GDIP01158781 GDIP01137338 GDIP01137337 GDIP01137282 GDIP01125277 GDIP01125231 GDIP01125230 GDIP01124181 GDIP01124180 GDIP01124179 GDIP01093038 GDIP01087383 GDIP01087382 GDIP01086288 GDIP01086287 GDIP01064250 GDIP01064249 GDIP01064248 GDIP01061339 GDIP01061338 GDIP01052294 GDIP01052293 GDIP01052292 JAJ63029.1 GDIP01180076 JAJ43326.1 GDIP01119429 JAL84285.1 GDIP01225466 JAI97935.1 GDIP01119428 JAL84286.1 GDIQ01080335 JAN14402.1 GDIP01239539 GDIP01238226 GDIP01236779 GDIP01231022 GDIP01227816 GDIP01225465 GDIP01222269 GDIP01222268 GDIP01220978 GDIP01210469 GDIP01201879 GDIP01200616 GDIP01197276 GDIP01177698 GDIP01137283 GDIP01125278 GDIP01124182 GDIP01093041 GDIP01087384 GDIP01086289 GDIP01064247 GDIP01061337 GDIP01061336 GDIP01052295 JAI85175.1 GDIP01121423 JAL82291.1 KQ414775 KOC61256.1 SSX17797.1 JXJN01007030 CH379070 KRT08548.1 KRT08551.1 JH432004 KRT08550.1 KA644640 AFP59269.1 CP012525 ALC43739.1 JRES01000375 KNC31851.1 EAL29979.2 CH933816 EDW17258.2 CH940647 EDW68707.2 JH816905 EKC33188.1 CH964101 EDW79495.1 CH902618 EDV40479.2 KPU78582.1 GDIQ01114627 JAL37099.1 AE014296 BT003798 AAN11560.2 AAO41481.1
AGBW02013750 OWR42879.1 RSAL01000027 RVE51926.1 ODYU01000891 SOQ36332.1 JTDY01007635 KOB65057.1 KQ971307 EFA11227.2 GEZM01028000 JAV86530.1 CH477471 EAT40408.1 GL764626 EFZ17426.1 GBYB01002284 GBYB01002287 GBYB01002289 GBYB01008691 JAG72051.1 JAG72054.1 JAG72056.1 JAG78458.1 KK107678 QOIP01000004 EZA48128.1 RLU24195.1 LBMM01004386 KMQ92477.1 KQ980874 KYN12049.1 KQ981810 KYN35441.1 KQ982907 KYQ49379.1 GL435171 EFN73868.1 KQ976500 KYM83011.1 ADTU01023603 GL888059 EGI68535.1 GANO01000473 JAB59398.1 KQ978068 KYM97414.1 GEBQ01029009 JAT10968.1 GECZ01019726 GECZ01009362 JAS50043.1 JAS60407.1 GECZ01003388 JAS66381.1 GECU01005987 JAT01720.1 KK852855 KDR14931.1 ACPB03021415 PYGN01000032 PSN56869.1 GBBI01002869 JAC15843.1 GFTR01008048 JAW08378.1 KQ435794 KOX73808.1 KZ288189 PBC34591.1 KK856447 PTY25774.1 GBHO01042284 GDHC01019872 JAG01320.1 JAP98756.1 KQ434827 KZC07616.1 UFQT01000016 SSX17796.1 CVRI01000042 CRK95487.1 DS231840 EDS34577.1 KQ767803 OAD53293.1 DS235201 EEB13226.1 AAAB01008964 EAA12917.4 APCN01002503 GDIQ01154211 JAK97514.1 GDIQ01055902 JAN38835.1 GL447585 EFN86353.1 GDIQ01058596 JAN36141.1 GDIQ01076035 JAN18702.1 GDIP01039643 JAM64072.1 GL732541 EFX82053.1 GDIP01235567 GDIP01229618 GDIP01196221 GDIP01195069 GDIP01191769 GDIP01185981 GDIP01181826 GDIP01181825 GDIP01180075 GDIP01180074 GDIP01154585 GDIP01146329 GDIQ01239492 GDIP01130987 GDIP01130986 GDIP01130985 GDIP01114239 GDIP01108129 GDIP01095893 GDIP01095892 GDIP01094690 GDIP01094689 GDIP01094688 GDIP01084991 GDIP01083245 GDIP01074392 GDIP01073145 GDIP01073144 GDIP01073095 GDIP01071684 GDIP01071683 GDIP01070142 GDIP01062993 GDIP01062992 GDIP01062991 GDIP01051047 GDIP01051046 GDIP01044050 GDIP01044049 GDIP01044048 GDIQ01081938 GDIQ01081937 GDIQ01035017 LRGB01002124 JAK12233.1 JAM18724.1 KZS08999.1 GDIQ01151311 JAL00415.1 GDIQ01194699 JAK57026.1 GDIP01229619 GDIP01195068 GDIP01195067 GDIP01191770 GDIP01185982 GDIP01181828 GDIP01180077 GDIP01177742 GDIP01146330 GDIP01130988 GDIP01114285 GDIP01114240 GDIP01110183 GDIP01108132 GDIP01105683 GDIP01095891 GDIP01094692 GDIP01084990 GDIP01083244 GDIP01073094 GDIP01071682 GDIP01070141 GDIP01062990 GDIP01044047 JAM32033.1 GDIP01105684 JAL98030.1 GDIP01225467 JAI97934.1 GDIP01167966 JAJ55436.1 GDIP01239540 GDIP01238228 GDIP01238227 GDIP01236780 GDIP01231025 GDIP01231024 GDIP01231023 GDIP01227817 GDIP01220980 GDIP01220979 GDIP01210470 GDIP01201877 GDIP01201876 GDIP01201875 GDIP01200619 GDIP01200618 GDIP01200617 GDIP01197278 GDIP01197277 GDIP01197274 GDIP01167023 GDIP01160373 GDIP01158781 GDIP01137338 GDIP01137337 GDIP01137282 GDIP01125277 GDIP01125231 GDIP01125230 GDIP01124181 GDIP01124180 GDIP01124179 GDIP01093038 GDIP01087383 GDIP01087382 GDIP01086288 GDIP01086287 GDIP01064250 GDIP01064249 GDIP01064248 GDIP01061339 GDIP01061338 GDIP01052294 GDIP01052293 GDIP01052292 JAJ63029.1 GDIP01180076 JAJ43326.1 GDIP01119429 JAL84285.1 GDIP01225466 JAI97935.1 GDIP01119428 JAL84286.1 GDIQ01080335 JAN14402.1 GDIP01239539 GDIP01238226 GDIP01236779 GDIP01231022 GDIP01227816 GDIP01225465 GDIP01222269 GDIP01222268 GDIP01220978 GDIP01210469 GDIP01201879 GDIP01200616 GDIP01197276 GDIP01177698 GDIP01137283 GDIP01125278 GDIP01124182 GDIP01093041 GDIP01087384 GDIP01086289 GDIP01064247 GDIP01061337 GDIP01061336 GDIP01052295 JAI85175.1 GDIP01121423 JAL82291.1 KQ414775 KOC61256.1 SSX17797.1 JXJN01007030 CH379070 KRT08548.1 KRT08551.1 JH432004 KRT08550.1 KA644640 AFP59269.1 CP012525 ALC43739.1 JRES01000375 KNC31851.1 EAL29979.2 CH933816 EDW17258.2 CH940647 EDW68707.2 JH816905 EKC33188.1 CH964101 EDW79495.1 CH902618 EDV40479.2 KPU78582.1 GDIQ01114627 JAL37099.1 AE014296 BT003798 AAN11560.2 AAO41481.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
UP000037510
+ More
UP000007266 UP000192223 UP000002358 UP000008820 UP000053097 UP000279307 UP000036403 UP000078492 UP000078541 UP000075809 UP000000311 UP000078540 UP000005205 UP000007755 UP000078542 UP000027135 UP000015103 UP000245037 UP000075880 UP000053105 UP000005203 UP000242457 UP000076502 UP000075885 UP000183832 UP000002320 UP000009046 UP000007062 UP000075840 UP000008237 UP000000305 UP000076858 UP000053825 UP000078200 UP000091820 UP000092445 UP000092443 UP000092460 UP000001819 UP000095300 UP000095301 UP000092553 UP000037069 UP000009192 UP000008792 UP000005408 UP000007798 UP000007801 UP000000803
UP000007266 UP000192223 UP000002358 UP000008820 UP000053097 UP000279307 UP000036403 UP000078492 UP000078541 UP000075809 UP000000311 UP000078540 UP000005205 UP000007755 UP000078542 UP000027135 UP000015103 UP000245037 UP000075880 UP000053105 UP000005203 UP000242457 UP000076502 UP000075885 UP000183832 UP000002320 UP000009046 UP000007062 UP000075840 UP000008237 UP000000305 UP000076858 UP000053825 UP000078200 UP000091820 UP000092445 UP000092443 UP000092460 UP000001819 UP000095300 UP000095301 UP000092553 UP000037069 UP000009192 UP000008792 UP000005408 UP000007798 UP000007801 UP000000803
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A194RNB1
A0A194PDG3
A0A2A4IY94
A0A212EN26
A0A3S2PHY4
A0A2H1V7H1
+ More
A0A0L7KP37 D6W7U6 A0A1W4X2N7 A0A1W4XDG2 A0A1Y1MPC4 K7IZ64 Q170P2 A0A1S4FHS4 A0A1S4J3C9 E9IPU7 A0A1S4J3S8 A0A0C9QNU7 A0A026VWI0 A0A0J7KQ46 A0A151IW48 A0A195F4X1 A0A151WNC6 E1ZY19 A0A195BEJ5 A0A158NQU1 F4WB84 U5EY75 A0A195C972 A0A1B6KHM4 A0A1B6GDB8 A0A1B6GVD4 A0A1B6JR58 A0A067R907 T1I949 A0A2P8ZK48 A0A182IZI2 A0A023F3F3 A0A224XF16 A0A0N0U536 A0A088A5L4 A0A2A3EU62 A0A2R7X1W1 A0A0A9W8Z8 A0A154P6P5 A0A336LIW2 A0A182PMD6 A0A1J1I572 B0W511 A0A310SDB1 E0VIM0 A0A1S4GXD8 Q7Q3J1 A0A182HVA7 A0A0N8BQQ6 A0A0P6G800 E2BDC5 A0A0N8E5Y4 A0A0P6EIG3 A0A0P6AQL9 E9GEM8 A0A0N8CZ07 A0A0P5MZI4 A0A0N8BB95 A0A0P5XII2 A0A0P5V3K0 A0A0P4YXY2 A0A0P5CR49 A0A0P5D5Z6 A0A0P5BTN9 A0A0P5U0P3 A0A0P4YQA0 A0A0P5U1L2 A0A0P6DA04 A0A0P4XQF4 A0A0N8CL03 A0A0L7QRX1 A0A336LN51 A0A1A9UJF2 A0A1A9W390 A0A1A9Z2P8 A0A1A9YEN6 A0A1B0B192 A0A0R3P5T1 T1JAT6 A0A0R3P914 A0A1I8Q8A8 T1P919 A0A0M3QW90 A0A0L0CHY1 Q29ET1 B4L983 B4LBT0 K1QH32 B4N556 B3M4I5 A0A0N8C4V4 Q8IRD5
A0A0L7KP37 D6W7U6 A0A1W4X2N7 A0A1W4XDG2 A0A1Y1MPC4 K7IZ64 Q170P2 A0A1S4FHS4 A0A1S4J3C9 E9IPU7 A0A1S4J3S8 A0A0C9QNU7 A0A026VWI0 A0A0J7KQ46 A0A151IW48 A0A195F4X1 A0A151WNC6 E1ZY19 A0A195BEJ5 A0A158NQU1 F4WB84 U5EY75 A0A195C972 A0A1B6KHM4 A0A1B6GDB8 A0A1B6GVD4 A0A1B6JR58 A0A067R907 T1I949 A0A2P8ZK48 A0A182IZI2 A0A023F3F3 A0A224XF16 A0A0N0U536 A0A088A5L4 A0A2A3EU62 A0A2R7X1W1 A0A0A9W8Z8 A0A154P6P5 A0A336LIW2 A0A182PMD6 A0A1J1I572 B0W511 A0A310SDB1 E0VIM0 A0A1S4GXD8 Q7Q3J1 A0A182HVA7 A0A0N8BQQ6 A0A0P6G800 E2BDC5 A0A0N8E5Y4 A0A0P6EIG3 A0A0P6AQL9 E9GEM8 A0A0N8CZ07 A0A0P5MZI4 A0A0N8BB95 A0A0P5XII2 A0A0P5V3K0 A0A0P4YXY2 A0A0P5CR49 A0A0P5D5Z6 A0A0P5BTN9 A0A0P5U0P3 A0A0P4YQA0 A0A0P5U1L2 A0A0P6DA04 A0A0P4XQF4 A0A0N8CL03 A0A0L7QRX1 A0A336LN51 A0A1A9UJF2 A0A1A9W390 A0A1A9Z2P8 A0A1A9YEN6 A0A1B0B192 A0A0R3P5T1 T1JAT6 A0A0R3P914 A0A1I8Q8A8 T1P919 A0A0M3QW90 A0A0L0CHY1 Q29ET1 B4L983 B4LBT0 K1QH32 B4N556 B3M4I5 A0A0N8C4V4 Q8IRD5
Ontologies
GO
PANTHER
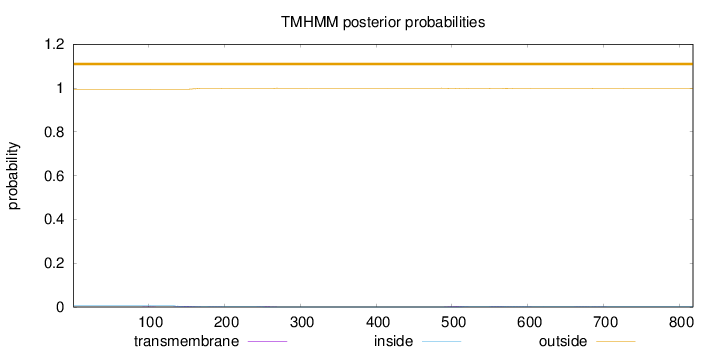
Topology
Length:
818
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23455
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00581
outside
1 - 818
Population Genetic Test Statistics
Pi
290.839181
Theta
232.116261
Tajima's D
0.730732
CLR
0.055635
CSRT
0.580570971451427
Interpretation
Uncertain
