Gene
KWMTBOMO05136
Pre Gene Modal
BGIBMGA012502
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_putative_sodium-dependent_multivitamin_transporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.972
Sequence
CDS
ATGGCGTTATCAGTATTTGGAATATGTGACTACCTTATAATGGTAGCGACTATGATCGCTTCTGTGGCTATTGGAGTCTATTTCCGATTTAGTGGAGGAAAACAGAAAACAAACGAGGAATACCTCTTAGCCGATAGAAACATGTCAATATTACCGGTCGCGGTTTCATTAATGGCCTCATTCATGTCGGCGATTACTCTTCTGGGAGTGTCGGCAGAAAACTACTATTTCGGCATGCAATTCGTTATCATAAATATAGCCTACTTCTTAGCAACTCCGATAGCTAGTAGATTATATCTACCCGTGTTCTTTGGCCTTCAAAAAACAAGTACCTATGAGTACTTAGAACTGAGATTTGGGTCTCAAATACGTATGCTGGCGTCTCTGACGTACACTTTACAAATGTTATTGTACAATGGCATAGTGCTTTATGCTCCAGCTATTGTATTAGAATCTGTGACTGGTTTAGACAGGATAATATCGATACTAGTTGTAGGCTTAGCTTGTACGTTTTACTCGACTTTAGGAGGGATGAAGGCCGTATTATTTACTGATTTACTGCAGTCTTTGCTGATGTTTGCCGCTGTGTTGAGTGTGGTGGTATTTAGCGCAGTCCAAGTCGGCGGATTCGGACAAATATTTGTTCGTGCCCGGGAAGGTGGACGATTAGACTTTTCCAATTTCAGTGTAGATCCCACCCAGAGGCACACTTGGTGGTCACTCATTATTGGAGGAATGATAACTTACTTATCACTGTATGCTGTCAATCATACTCAGGTACAACGTCTGTTAACAGTGAGCACGTTGGACCGATCCCAGCAGTGCCTCTGGTGGAATTTGCCCGTGATGTCTGCTCTCAGTATCATTACATGCATCAGTGGATTAGGCATCTACGCCGTCTATAAAGACTGTGATCCGTTGACTTATCACGGAATTACTGCTATTGACCAACTGATGCCTTACTACGTGGTACAAATGATGAAGTCAGCTCCCGGTCTTCTAGGATTATTCATCGCTGGTATATTCAGTGCTTCCCTCTCCACGATATCAGCGGCTTGTAATGCACTGGCCGCAGTGACTCTAACTGATTACGTCAACAGATGGTTCTTAATACCTTCATCCTATATACCCTGGATGACAAAGCTAGCAGCCTGTACTTATGGATTCCTATTCCTCGGCCTTGCTTTCTTGGCTGAACATTTAGGAGGGGTATTGCAAGCAGCTTTGACTATCTTTGGAGCTGTTGGGGGCCCGTTGTTCGGAGTTTTTACTTTAGGCATGTTTACTACTTTTGCAAATGAGAGGGGCGCATTCGTCGCTCTATTAACTGGCATGGCGCTTACTCTGTGGATGAGTTTCGGCGGACCGCGACCGACGCCTGCGAAACTGCCGCTAAGCGTAGACGGATGCGGTTTTAATGTCACCAATCCACTACCGGCCGTGTCGACGAACTTCAGTGACTACTTTTACCCGTACAGAATATCGTACATGTGGACCAGTCCGATTGGATTCGTCTGGGTGTTAGCTGTTGGGACAGCGATATCTTTAGTGTGGCGACAGCAACAACCATGGGAACGCGATGGGAAACCCCATCCTGACCCCGCGCTCTTCACTCCGCCCCTTGCCAAGCTACTTAGAAAGAAATATGGTGATAAGCCTCCCGTACAGCGCGGATTATTGAAGGAATCAAGTCTACATAATTAA
Protein
MALSVFGICDYLIMVATMIASVAIGVYFRFSGGKQKTNEEYLLADRNMSILPVAVSLMASFMSAITLLGVSAENYYFGMQFVIINIAYFLATPIASRLYLPVFFGLQKTSTYEYLELRFGSQIRMLASLTYTLQMLLYNGIVLYAPAIVLESVTGLDRIISILVVGLACTFYSTLGGMKAVLFTDLLQSLLMFAAVLSVVVFSAVQVGGFGQIFVRAREGGRLDFSNFSVDPTQRHTWWSLIIGGMITYLSLYAVNHTQVQRLLTVSTLDRSQQCLWWNLPVMSALSIITCISGLGIYAVYKDCDPLTYHGITAIDQLMPYYVVQMMKSAPGLLGLFIAGIFSASLSTISAACNALAAVTLTDYVNRWFLIPSSYIPWMTKLAACTYGFLFLGLAFLAEHLGGVLQAALTIFGAVGGPLFGVFTLGMFTTFANERGAFVALLTGMALTLWMSFGGPRPTPAKLPLSVDGCGFNVTNPLPAVSTNFSDYFYPYRISYMWTSPIGFVWVLAVGTAISLVWRQQQPWERDGKPHPDPALFTPPLAKLLRKKYGDKPPVQRGLLKESSLHN
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A2A4J858
A0A2W1BLM0
A0A2A4J950
A0A194PDF8
A0A437BNF5
A0A2H1W3B3
+ More
A0A194PJV6 A0A0L7LLE5 A0A194RMZ9 A0A1Y1K0U1 A0A1Y1K0W9 A0A1Y1K127 A0A1Y1K0V3 J3JWA1 N6TW03 N6T083 W8BLK9 A0A182W1E8 A0A182RXP5 D6WJW4 A0A182YGR2 A0A182IJ72 A0A182NLF2 A0A182PKP7 U5EW06 A0A182Q3I9 U5EVD2 A0A1L8DF14 A0A2M4BHR8 W5JBY9 A0A2M4AFP2 A0A182V5C8 A0A2M4BHU7 Q7QB40 A0A182FSB5 A0A182UEH1 A0A0A1XGG9 A0A182M8Q9 A0A1S4FCK0 A0A1I8M662 A0A1Q3FDK3 Q177I0 A0A1Q3FBD4 A0A0K8WMM0 A0A1Q3FDF5 B0WU59 A0A182XJ67 A0A182I7P3 A0A182JQ24 A0A1I8PQL8 E9GP61 A0A026X1Y2 Q8I923 A0A182H517 A0A232F6S4 A0A0N8CW20 A0A0P5QMF0 U5EVE6 A0A0P6E8F0 A0A182T9P8 A0A0N8DXH3 A0A0L7R0H8 A0A0P5SZS6 A0A195EY23 A0A0P5N8J5 A0A0P5W478 A0A0P5XC05 A0A023EX78 A0A0P6FMK5 A0A1A9ZED7 A0A1A9XLM6 A0A0P5WZX6 A0A1L8ECI2 A0A1L8EC33 A0A1L8EC39 A0A067RCW7 B4PAN2 F4X3J0 A0A2M4AGL8 A0A1A9W305 A0A1B0B6B1 A0A336M4V5 A0A158NRN4 Q5R281 Q5R268 A0A1A9VUD5 A0A154NY42 B4LPN3 B3NK51 A0A0P6EQF8 A0A1W4V4U7 A0A0P5AG34 Q8SWV5 A0A087ZNV1 A0A151X8Z7 B4QEL6 A0A2A3EQG2 A0A0M8ZXX9 A0A1B6C0C0
A0A194PJV6 A0A0L7LLE5 A0A194RMZ9 A0A1Y1K0U1 A0A1Y1K0W9 A0A1Y1K127 A0A1Y1K0V3 J3JWA1 N6TW03 N6T083 W8BLK9 A0A182W1E8 A0A182RXP5 D6WJW4 A0A182YGR2 A0A182IJ72 A0A182NLF2 A0A182PKP7 U5EW06 A0A182Q3I9 U5EVD2 A0A1L8DF14 A0A2M4BHR8 W5JBY9 A0A2M4AFP2 A0A182V5C8 A0A2M4BHU7 Q7QB40 A0A182FSB5 A0A182UEH1 A0A0A1XGG9 A0A182M8Q9 A0A1S4FCK0 A0A1I8M662 A0A1Q3FDK3 Q177I0 A0A1Q3FBD4 A0A0K8WMM0 A0A1Q3FDF5 B0WU59 A0A182XJ67 A0A182I7P3 A0A182JQ24 A0A1I8PQL8 E9GP61 A0A026X1Y2 Q8I923 A0A182H517 A0A232F6S4 A0A0N8CW20 A0A0P5QMF0 U5EVE6 A0A0P6E8F0 A0A182T9P8 A0A0N8DXH3 A0A0L7R0H8 A0A0P5SZS6 A0A195EY23 A0A0P5N8J5 A0A0P5W478 A0A0P5XC05 A0A023EX78 A0A0P6FMK5 A0A1A9ZED7 A0A1A9XLM6 A0A0P5WZX6 A0A1L8ECI2 A0A1L8EC33 A0A1L8EC39 A0A067RCW7 B4PAN2 F4X3J0 A0A2M4AGL8 A0A1A9W305 A0A1B0B6B1 A0A336M4V5 A0A158NRN4 Q5R281 Q5R268 A0A1A9VUD5 A0A154NY42 B4LPN3 B3NK51 A0A0P6EQF8 A0A1W4V4U7 A0A0P5AG34 Q8SWV5 A0A087ZNV1 A0A151X8Z7 B4QEL6 A0A2A3EQG2 A0A0M8ZXX9 A0A1B6C0C0
Pubmed
28756777
26354079
26227816
28004739
22516182
23537049
+ More
24495485 18362917 19820115 25244985 20920257 23761445 12364791 14747013 17210077 25830018 25315136 17510324 21292972 24508170 30249741 26483478 28648823 24945155 24845553 17994087 17550304 21719571 21347285 15729003 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
24495485 18362917 19820115 25244985 20920257 23761445 12364791 14747013 17210077 25830018 25315136 17510324 21292972 24508170 30249741 26483478 28648823 24945155 24845553 17994087 17550304 21719571 21347285 15729003 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
EMBL
NWSH01002573
PCG67946.1
KZ149974
PZC75982.1
PCG67942.1
KQ459606
+ More
KPI91376.1 RSAL01000027 RVE51931.1 ODYU01006040 SOQ47527.1 KPI91375.1 JTDY01000637 KOB76383.1 KQ459984 KPJ18897.1 GEZM01096160 GEZM01096152 GEZM01096149 JAV55122.1 GEZM01096155 JAV55119.1 GEZM01096151 JAV55123.1 GEZM01096153 GEZM01096150 JAV55124.1 BT127519 AEE62481.1 APGK01049912 APGK01049913 APGK01049914 APGK01049915 KB741159 ENN73470.1 ENN73469.1 GAMC01016021 JAB90534.1 KQ971342 EFA03653.1 GANO01001653 JAB58218.1 AXCN02000131 GANO01001909 JAB57962.1 GFDF01009109 JAV04975.1 GGFJ01003392 MBW52533.1 ADMH02001952 ETN60334.1 GGFK01006262 MBW39583.1 GGFJ01003422 MBW52563.1 AAAB01008880 EAA08654.4 GBXI01015079 GBXI01004694 JAC99212.1 JAD09598.1 AXCM01001304 GFDL01009473 JAV25572.1 CH477376 EAT42297.1 GFDL01010164 JAV24881.1 GDHF01000164 JAI52150.1 GFDL01009513 JAV25532.1 DS232101 EDS34779.1 APCN01002523 GL732556 EFX78734.1 KK107021 QOIP01000011 EZA62320.1 RLU16562.1 AY063742 AAL38977.1 JXUM01110571 KQ565425 KXJ70994.1 NNAY01000868 OXU26138.1 GDIP01093287 JAM10428.1 GDIQ01211898 GDIQ01122120 GDIQ01075653 GDIQ01039813 JAL29606.1 GANO01001911 JAB57960.1 GDIQ01066346 JAN28391.1 GDIQ01082284 JAN12453.1 KQ414670 KOC64350.1 GDIP01133032 JAL70682.1 KQ981931 KYN32797.1 GDIQ01145737 JAL05989.1 GDIP01091632 JAM12083.1 GDIP01074397 JAM29318.1 GAPW01000609 JAC12989.1 GDIQ01057302 JAN37435.1 GDIP01079836 JAM23879.1 GFDG01002490 JAV16309.1 GFDG01002491 JAV16308.1 GFDG01002489 JAV16310.1 KK852543 KDR21691.1 CM000158 EDW91423.1 GL888613 EGI59018.1 GGFK01006610 MBW39931.1 JXJN01009045 UFQS01000397 UFQT01000397 SSX03608.1 SSX23973.1 ADTU01024212 ADTU01024213 AB162350 BAD72905.1 AB162351 CH480816 BAD72923.1 EDW48677.1 KQ434781 KZC04501.1 CH940648 EDW60271.1 CH954179 EDV55286.1 GDIQ01059850 JAN34887.1 GDIP01199624 JAJ23778.1 AE013599 AY095060 AAF57489.2 AAM11388.1 KQ982409 KYQ56790.1 CM000362 CM002911 EDX07890.1 KMY95235.1 KZ288204 PBC33281.1 KQ435798 KOX73365.1 GEDC01030350 JAS06948.1
KPI91376.1 RSAL01000027 RVE51931.1 ODYU01006040 SOQ47527.1 KPI91375.1 JTDY01000637 KOB76383.1 KQ459984 KPJ18897.1 GEZM01096160 GEZM01096152 GEZM01096149 JAV55122.1 GEZM01096155 JAV55119.1 GEZM01096151 JAV55123.1 GEZM01096153 GEZM01096150 JAV55124.1 BT127519 AEE62481.1 APGK01049912 APGK01049913 APGK01049914 APGK01049915 KB741159 ENN73470.1 ENN73469.1 GAMC01016021 JAB90534.1 KQ971342 EFA03653.1 GANO01001653 JAB58218.1 AXCN02000131 GANO01001909 JAB57962.1 GFDF01009109 JAV04975.1 GGFJ01003392 MBW52533.1 ADMH02001952 ETN60334.1 GGFK01006262 MBW39583.1 GGFJ01003422 MBW52563.1 AAAB01008880 EAA08654.4 GBXI01015079 GBXI01004694 JAC99212.1 JAD09598.1 AXCM01001304 GFDL01009473 JAV25572.1 CH477376 EAT42297.1 GFDL01010164 JAV24881.1 GDHF01000164 JAI52150.1 GFDL01009513 JAV25532.1 DS232101 EDS34779.1 APCN01002523 GL732556 EFX78734.1 KK107021 QOIP01000011 EZA62320.1 RLU16562.1 AY063742 AAL38977.1 JXUM01110571 KQ565425 KXJ70994.1 NNAY01000868 OXU26138.1 GDIP01093287 JAM10428.1 GDIQ01211898 GDIQ01122120 GDIQ01075653 GDIQ01039813 JAL29606.1 GANO01001911 JAB57960.1 GDIQ01066346 JAN28391.1 GDIQ01082284 JAN12453.1 KQ414670 KOC64350.1 GDIP01133032 JAL70682.1 KQ981931 KYN32797.1 GDIQ01145737 JAL05989.1 GDIP01091632 JAM12083.1 GDIP01074397 JAM29318.1 GAPW01000609 JAC12989.1 GDIQ01057302 JAN37435.1 GDIP01079836 JAM23879.1 GFDG01002490 JAV16309.1 GFDG01002491 JAV16308.1 GFDG01002489 JAV16310.1 KK852543 KDR21691.1 CM000158 EDW91423.1 GL888613 EGI59018.1 GGFK01006610 MBW39931.1 JXJN01009045 UFQS01000397 UFQT01000397 SSX03608.1 SSX23973.1 ADTU01024212 ADTU01024213 AB162350 BAD72905.1 AB162351 CH480816 BAD72923.1 EDW48677.1 KQ434781 KZC04501.1 CH940648 EDW60271.1 CH954179 EDV55286.1 GDIQ01059850 JAN34887.1 GDIP01199624 JAJ23778.1 AE013599 AY095060 AAF57489.2 AAM11388.1 KQ982409 KYQ56790.1 CM000362 CM002911 EDX07890.1 KMY95235.1 KZ288204 PBC33281.1 KQ435798 KOX73365.1 GEDC01030350 JAS06948.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000037510
UP000053240
UP000019118
+ More
UP000075920 UP000075900 UP000007266 UP000076408 UP000075880 UP000075884 UP000075885 UP000075886 UP000000673 UP000075903 UP000007062 UP000069272 UP000075902 UP000075883 UP000095301 UP000008820 UP000002320 UP000076407 UP000075840 UP000075881 UP000095300 UP000000305 UP000053097 UP000279307 UP000069940 UP000249989 UP000215335 UP000075901 UP000053825 UP000078541 UP000092445 UP000092443 UP000027135 UP000002282 UP000007755 UP000091820 UP000092460 UP000005205 UP000001292 UP000078200 UP000076502 UP000008792 UP000008711 UP000192221 UP000000803 UP000005203 UP000075809 UP000000304 UP000242457 UP000053105
UP000075920 UP000075900 UP000007266 UP000076408 UP000075880 UP000075884 UP000075885 UP000075886 UP000000673 UP000075903 UP000007062 UP000069272 UP000075902 UP000075883 UP000095301 UP000008820 UP000002320 UP000076407 UP000075840 UP000075881 UP000095300 UP000000305 UP000053097 UP000279307 UP000069940 UP000249989 UP000215335 UP000075901 UP000053825 UP000078541 UP000092445 UP000092443 UP000027135 UP000002282 UP000007755 UP000091820 UP000092460 UP000005205 UP000001292 UP000078200 UP000076502 UP000008792 UP000008711 UP000192221 UP000000803 UP000005203 UP000075809 UP000000304 UP000242457 UP000053105
PRIDE
Pfam
PF00474 SSF
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J858
A0A2W1BLM0
A0A2A4J950
A0A194PDF8
A0A437BNF5
A0A2H1W3B3
+ More
A0A194PJV6 A0A0L7LLE5 A0A194RMZ9 A0A1Y1K0U1 A0A1Y1K0W9 A0A1Y1K127 A0A1Y1K0V3 J3JWA1 N6TW03 N6T083 W8BLK9 A0A182W1E8 A0A182RXP5 D6WJW4 A0A182YGR2 A0A182IJ72 A0A182NLF2 A0A182PKP7 U5EW06 A0A182Q3I9 U5EVD2 A0A1L8DF14 A0A2M4BHR8 W5JBY9 A0A2M4AFP2 A0A182V5C8 A0A2M4BHU7 Q7QB40 A0A182FSB5 A0A182UEH1 A0A0A1XGG9 A0A182M8Q9 A0A1S4FCK0 A0A1I8M662 A0A1Q3FDK3 Q177I0 A0A1Q3FBD4 A0A0K8WMM0 A0A1Q3FDF5 B0WU59 A0A182XJ67 A0A182I7P3 A0A182JQ24 A0A1I8PQL8 E9GP61 A0A026X1Y2 Q8I923 A0A182H517 A0A232F6S4 A0A0N8CW20 A0A0P5QMF0 U5EVE6 A0A0P6E8F0 A0A182T9P8 A0A0N8DXH3 A0A0L7R0H8 A0A0P5SZS6 A0A195EY23 A0A0P5N8J5 A0A0P5W478 A0A0P5XC05 A0A023EX78 A0A0P6FMK5 A0A1A9ZED7 A0A1A9XLM6 A0A0P5WZX6 A0A1L8ECI2 A0A1L8EC33 A0A1L8EC39 A0A067RCW7 B4PAN2 F4X3J0 A0A2M4AGL8 A0A1A9W305 A0A1B0B6B1 A0A336M4V5 A0A158NRN4 Q5R281 Q5R268 A0A1A9VUD5 A0A154NY42 B4LPN3 B3NK51 A0A0P6EQF8 A0A1W4V4U7 A0A0P5AG34 Q8SWV5 A0A087ZNV1 A0A151X8Z7 B4QEL6 A0A2A3EQG2 A0A0M8ZXX9 A0A1B6C0C0
A0A194PJV6 A0A0L7LLE5 A0A194RMZ9 A0A1Y1K0U1 A0A1Y1K0W9 A0A1Y1K127 A0A1Y1K0V3 J3JWA1 N6TW03 N6T083 W8BLK9 A0A182W1E8 A0A182RXP5 D6WJW4 A0A182YGR2 A0A182IJ72 A0A182NLF2 A0A182PKP7 U5EW06 A0A182Q3I9 U5EVD2 A0A1L8DF14 A0A2M4BHR8 W5JBY9 A0A2M4AFP2 A0A182V5C8 A0A2M4BHU7 Q7QB40 A0A182FSB5 A0A182UEH1 A0A0A1XGG9 A0A182M8Q9 A0A1S4FCK0 A0A1I8M662 A0A1Q3FDK3 Q177I0 A0A1Q3FBD4 A0A0K8WMM0 A0A1Q3FDF5 B0WU59 A0A182XJ67 A0A182I7P3 A0A182JQ24 A0A1I8PQL8 E9GP61 A0A026X1Y2 Q8I923 A0A182H517 A0A232F6S4 A0A0N8CW20 A0A0P5QMF0 U5EVE6 A0A0P6E8F0 A0A182T9P8 A0A0N8DXH3 A0A0L7R0H8 A0A0P5SZS6 A0A195EY23 A0A0P5N8J5 A0A0P5W478 A0A0P5XC05 A0A023EX78 A0A0P6FMK5 A0A1A9ZED7 A0A1A9XLM6 A0A0P5WZX6 A0A1L8ECI2 A0A1L8EC33 A0A1L8EC39 A0A067RCW7 B4PAN2 F4X3J0 A0A2M4AGL8 A0A1A9W305 A0A1B0B6B1 A0A336M4V5 A0A158NRN4 Q5R281 Q5R268 A0A1A9VUD5 A0A154NY42 B4LPN3 B3NK51 A0A0P6EQF8 A0A1W4V4U7 A0A0P5AG34 Q8SWV5 A0A087ZNV1 A0A151X8Z7 B4QEL6 A0A2A3EQG2 A0A0M8ZXX9 A0A1B6C0C0
PDB
5NVA
E-value=2.75777e-21,
Score=253
Ontologies
GO
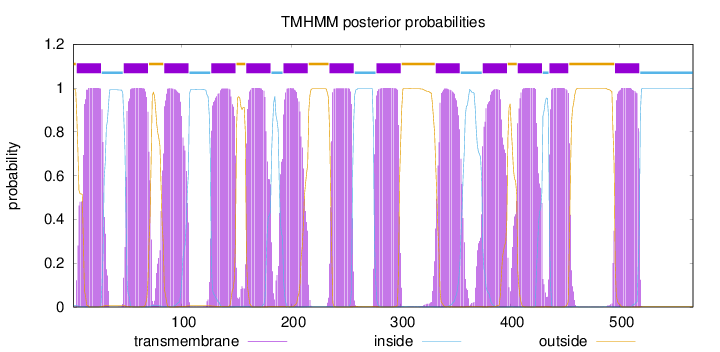
Topology
Length:
567
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
287.725570000001
Exp number, first 60 AAs:
33.43928
Total prob of N-in:
0.00829
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 46
TMhelix
47 - 69
outside
70 - 83
TMhelix
84 - 106
inside
107 - 126
TMhelix
127 - 149
outside
150 - 158
TMhelix
159 - 181
inside
182 - 192
TMhelix
193 - 215
outside
216 - 234
TMhelix
235 - 257
inside
258 - 277
TMhelix
278 - 300
outside
301 - 331
TMhelix
332 - 354
inside
355 - 374
TMhelix
375 - 397
outside
398 - 406
TMhelix
407 - 429
inside
430 - 435
TMhelix
436 - 453
outside
454 - 495
TMhelix
496 - 518
inside
519 - 567
Population Genetic Test Statistics
Pi
215.493594
Theta
172.473944
Tajima's D
1.234378
CLR
0.150846
CSRT
0.723813809309535
Interpretation
Uncertain
