Gene
KWMTBOMO05126
Annotation
PREDICTED:_migration_and_invasion_enhancer_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.916
Sequence
CDS
ATGACTGGGAATGAAGAAAAAAGTTTCATCCCTAAAGTACATGTAGAATACTGTGGTGCTTGTGACTATGGAGGTCATTGCTTAGCCCTGGCGCAAATTATTAGAAATGATACACCTTCTGCTATGGTTTTGTGTAAAAGGGGACGTCAAGGGTCTTTTGAAGTAGTTGTGAATGATAAGTTGATTTACTCTAAGCTCAAAACAATGGCATTGCCAGATTATGGAGAAGTAGCTCAAGTAGTTCACGATGTTTCAAAAGGCCAAGAACCAAGGGAAATCAAAGGAGAACAACCAATCAATTGTGCTATATCTTGA
Protein
MTGNEEKSFIPKVHVEYCGACDYGGHCLALAQIIRNDTPSAMVLCKRGRQGSFEVVVNDKLIYSKLKTMALPDYGEVAQVVHDVSKGQEPREIKGEQPINCAIS
Summary
Uniprot
S4P0M0
A0A2A4J1P7
A0A2H1VA03
A0A212FC68
A0A194RM36
A0A0J7LBG7
+ More
A0A0K8TNU5 A0A182W4P6 A0A2P8XN98 A0A158NDB9 A0A2M4B064 A0A182M1C4 A0A182FK20 A0A2M3ZDR0 A0A2M3ZKG9 A0A084VVL3 W5JL32 A0A2M4C4H7 A0A2M4AZK6 A0A182K4H6 A0A3L8DRU5 A0A182P7X3 A0A1I8P4F5 A0A182XLS0 A0A182IA79 A0A182V3Z8 A0A0L0C241 A0A182Y7I8 A0A1Q3F8E7 A0A023ED07 A0A2M4AZL6 A0A182G7C5 A0A1S4FUI2 Q16P89 A0A1J1HHR8 A0A1I8MC84 B0WSX0 A0A034WNF4 A0A023EEN1 A7L483 A0A023ED31 A0A232ELY3 A0A1B6CDG1 K7J4W5 A0A151WES7 A0A1L8E4I4 B3MZN5 B4PYH3 A0A1L8E460 A0A182TZF6 B4I683 A0A1W4VT44 B4M3C7 U4UUM6 Q9VRA0 B3NWJ7 D6WBT4 A0A195FI18 B7FNM1 B4JXP9 A0A0K8UMT8 Q7QFL8 A0A182LGY5 B4L338 A0A034WMK6 A0A0P6HZV7 B4NDQ5 Q16GH1 V9L4J1 A0A182N1H6 Q29GM1 A0A195C380 B4H0T1 A0A0M3QZG9 A0A1A9XXP9 A0A3R7PJ90 A0A1L8E4C7 A0A1W4ZNP3 F6NX26 A0A2I4CCC3 A0A1B6HJK7 Q3ZLC7 Q802F5 A0A2D0T2A6 A0A2K5IB97 A0A131XPR8 K4G7U2 V9LHW2 A0A2K5KRG9 A0A2R5LBD8 A0A2K5XJK8 G7PUM5
A0A0K8TNU5 A0A182W4P6 A0A2P8XN98 A0A158NDB9 A0A2M4B064 A0A182M1C4 A0A182FK20 A0A2M3ZDR0 A0A2M3ZKG9 A0A084VVL3 W5JL32 A0A2M4C4H7 A0A2M4AZK6 A0A182K4H6 A0A3L8DRU5 A0A182P7X3 A0A1I8P4F5 A0A182XLS0 A0A182IA79 A0A182V3Z8 A0A0L0C241 A0A182Y7I8 A0A1Q3F8E7 A0A023ED07 A0A2M4AZL6 A0A182G7C5 A0A1S4FUI2 Q16P89 A0A1J1HHR8 A0A1I8MC84 B0WSX0 A0A034WNF4 A0A023EEN1 A7L483 A0A023ED31 A0A232ELY3 A0A1B6CDG1 K7J4W5 A0A151WES7 A0A1L8E4I4 B3MZN5 B4PYH3 A0A1L8E460 A0A182TZF6 B4I683 A0A1W4VT44 B4M3C7 U4UUM6 Q9VRA0 B3NWJ7 D6WBT4 A0A195FI18 B7FNM1 B4JXP9 A0A0K8UMT8 Q7QFL8 A0A182LGY5 B4L338 A0A034WMK6 A0A0P6HZV7 B4NDQ5 Q16GH1 V9L4J1 A0A182N1H6 Q29GM1 A0A195C380 B4H0T1 A0A0M3QZG9 A0A1A9XXP9 A0A3R7PJ90 A0A1L8E4C7 A0A1W4ZNP3 F6NX26 A0A2I4CCC3 A0A1B6HJK7 Q3ZLC7 Q802F5 A0A2D0T2A6 A0A2K5IB97 A0A131XPR8 K4G7U2 V9LHW2 A0A2K5KRG9 A0A2R5LBD8 A0A2K5XJK8 G7PUM5
Pubmed
23622113
22118469
26354079
26369729
29403074
21347285
+ More
24438588 20920257 23761445 30249741 26108605 25244985 24945155 26483478 17510324 25315136 25348373 28648823 20075255 17994087 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 12364791 20966253 24402279 15632085 23594743 12915322 28049606 23056606 22002653
24438588 20920257 23761445 30249741 26108605 25244985 24945155 26483478 17510324 25315136 25348373 28648823 20075255 17994087 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 12364791 20966253 24402279 15632085 23594743 12915322 28049606 23056606 22002653
EMBL
GAIX01009276
JAA83284.1
NWSH01003697
PCG65911.1
ODYU01001457
SOQ37680.1
+ More
AGBW02009211 OWR51339.1 KQ459984 KPJ18888.1 LBMM01000043 KMR05183.1 GDAI01001785 JAI15818.1 PYGN01001670 PSN33466.1 ADTU01000165 GGFK01012887 MBW46208.1 AXCM01014823 GGFM01005933 MBW26684.1 GGFM01008224 MBW28975.1 ATLV01017197 KE525157 KFB42007.1 ADMH02001131 ETN64008.1 GGFJ01011082 MBW60223.1 GGFK01012912 MBW46233.1 QOIP01000005 RLU23150.1 APCN01001132 JRES01000996 KNC26336.1 GFDL01011195 JAV23850.1 GAPW01006300 JAC07298.1 GGFK01012915 MBW46236.1 JXUM01046438 KQ561477 KXJ78467.1 CH477791 EAT36179.1 CVRI01000004 CRK87435.1 DS232077 EDS34098.1 GAKP01003070 GAKP01003067 GAKP01003065 JAC55882.1 GAPW01006302 JAC07296.1 EF660893 ABS19961.1 GAPW01006301 JAC07297.1 NNAY01003469 OXU19356.1 GEDC01025988 JAS11310.1 AAZX01000454 KQ983238 KYQ46306.1 GFDF01000540 JAV13544.1 CH902635 EDV33836.1 CM000162 EDX03014.1 GFDF01000638 JAV13446.1 CH480823 EDW56289.1 CH940651 EDW65302.1 KB632375 ERL93891.1 AE014298 AAF50903.1 AHN59961.1 CH954180 EDV46817.1 KQ971314 EEZ98745.1 KQ981606 KYN39634.1 BT053711 ACK77629.1 CH916376 EDV95148.1 GDHF01024516 JAI27798.1 AAAB01008846 EAA06627.4 CH933810 EDW06966.1 GAKP01003068 JAC55884.1 GDIQ01019505 JAN75232.1 CH964239 EDW81877.1 CH478278 EAT33328.1 JW873548 AFP06065.1 CH379064 EAL32088.1 KQ978350 KYM94651.1 CH479201 EDW29996.1 CP012528 ALC49403.1 QCYY01002014 ROT73615.1 GFDF01000568 JAV13516.1 CT573444 GECU01032837 JAS74869.1 AY737049 AAX61159.1 BC162535 BC162538 AY221261 AAI62535.1 AAO65270.1 GEFH01000229 JAP68352.1 JX209307 JX209470 JX209595 JX209603 JX209794 JX209795 JX209987 JX210336 JX210549 JX210649 JX210889 JX211066 JX211082 JX211122 JX211188 JW879663 AFM87621.1 AFM88301.1 AFP12180.1 JW879710 AFP12227.1 GGLE01002541 MBY06667.1 CM001291 EHH58074.1
AGBW02009211 OWR51339.1 KQ459984 KPJ18888.1 LBMM01000043 KMR05183.1 GDAI01001785 JAI15818.1 PYGN01001670 PSN33466.1 ADTU01000165 GGFK01012887 MBW46208.1 AXCM01014823 GGFM01005933 MBW26684.1 GGFM01008224 MBW28975.1 ATLV01017197 KE525157 KFB42007.1 ADMH02001131 ETN64008.1 GGFJ01011082 MBW60223.1 GGFK01012912 MBW46233.1 QOIP01000005 RLU23150.1 APCN01001132 JRES01000996 KNC26336.1 GFDL01011195 JAV23850.1 GAPW01006300 JAC07298.1 GGFK01012915 MBW46236.1 JXUM01046438 KQ561477 KXJ78467.1 CH477791 EAT36179.1 CVRI01000004 CRK87435.1 DS232077 EDS34098.1 GAKP01003070 GAKP01003067 GAKP01003065 JAC55882.1 GAPW01006302 JAC07296.1 EF660893 ABS19961.1 GAPW01006301 JAC07297.1 NNAY01003469 OXU19356.1 GEDC01025988 JAS11310.1 AAZX01000454 KQ983238 KYQ46306.1 GFDF01000540 JAV13544.1 CH902635 EDV33836.1 CM000162 EDX03014.1 GFDF01000638 JAV13446.1 CH480823 EDW56289.1 CH940651 EDW65302.1 KB632375 ERL93891.1 AE014298 AAF50903.1 AHN59961.1 CH954180 EDV46817.1 KQ971314 EEZ98745.1 KQ981606 KYN39634.1 BT053711 ACK77629.1 CH916376 EDV95148.1 GDHF01024516 JAI27798.1 AAAB01008846 EAA06627.4 CH933810 EDW06966.1 GAKP01003068 JAC55884.1 GDIQ01019505 JAN75232.1 CH964239 EDW81877.1 CH478278 EAT33328.1 JW873548 AFP06065.1 CH379064 EAL32088.1 KQ978350 KYM94651.1 CH479201 EDW29996.1 CP012528 ALC49403.1 QCYY01002014 ROT73615.1 GFDF01000568 JAV13516.1 CT573444 GECU01032837 JAS74869.1 AY737049 AAX61159.1 BC162535 BC162538 AY221261 AAI62535.1 AAO65270.1 GEFH01000229 JAP68352.1 JX209307 JX209470 JX209595 JX209603 JX209794 JX209795 JX209987 JX210336 JX210549 JX210649 JX210889 JX211066 JX211082 JX211122 JX211188 JW879663 AFM87621.1 AFM88301.1 AFP12180.1 JW879710 AFP12227.1 GGLE01002541 MBY06667.1 CM001291 EHH58074.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000036403
UP000075920
UP000245037
+ More
UP000005205 UP000075883 UP000069272 UP000030765 UP000000673 UP000075881 UP000279307 UP000075885 UP000095300 UP000076407 UP000075840 UP000075903 UP000037069 UP000076408 UP000069940 UP000249989 UP000008820 UP000183832 UP000095301 UP000002320 UP000215335 UP000002358 UP000075809 UP000007801 UP000002282 UP000075902 UP000001292 UP000192221 UP000008792 UP000030742 UP000000803 UP000008711 UP000007266 UP000078541 UP000001070 UP000007062 UP000075882 UP000009192 UP000007798 UP000075884 UP000001819 UP000078542 UP000008744 UP000092553 UP000092443 UP000283509 UP000192224 UP000000437 UP000192220 UP000221080 UP000233080 UP000233060 UP000233140 UP000009130
UP000005205 UP000075883 UP000069272 UP000030765 UP000000673 UP000075881 UP000279307 UP000075885 UP000095300 UP000076407 UP000075840 UP000075903 UP000037069 UP000076408 UP000069940 UP000249989 UP000008820 UP000183832 UP000095301 UP000002320 UP000215335 UP000002358 UP000075809 UP000007801 UP000002282 UP000075902 UP000001292 UP000192221 UP000008792 UP000030742 UP000000803 UP000008711 UP000007266 UP000078541 UP000001070 UP000007062 UP000075882 UP000009192 UP000007798 UP000075884 UP000001819 UP000078542 UP000008744 UP000092553 UP000092443 UP000283509 UP000192224 UP000000437 UP000192220 UP000221080 UP000233080 UP000233060 UP000233140 UP000009130
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
S4P0M0
A0A2A4J1P7
A0A2H1VA03
A0A212FC68
A0A194RM36
A0A0J7LBG7
+ More
A0A0K8TNU5 A0A182W4P6 A0A2P8XN98 A0A158NDB9 A0A2M4B064 A0A182M1C4 A0A182FK20 A0A2M3ZDR0 A0A2M3ZKG9 A0A084VVL3 W5JL32 A0A2M4C4H7 A0A2M4AZK6 A0A182K4H6 A0A3L8DRU5 A0A182P7X3 A0A1I8P4F5 A0A182XLS0 A0A182IA79 A0A182V3Z8 A0A0L0C241 A0A182Y7I8 A0A1Q3F8E7 A0A023ED07 A0A2M4AZL6 A0A182G7C5 A0A1S4FUI2 Q16P89 A0A1J1HHR8 A0A1I8MC84 B0WSX0 A0A034WNF4 A0A023EEN1 A7L483 A0A023ED31 A0A232ELY3 A0A1B6CDG1 K7J4W5 A0A151WES7 A0A1L8E4I4 B3MZN5 B4PYH3 A0A1L8E460 A0A182TZF6 B4I683 A0A1W4VT44 B4M3C7 U4UUM6 Q9VRA0 B3NWJ7 D6WBT4 A0A195FI18 B7FNM1 B4JXP9 A0A0K8UMT8 Q7QFL8 A0A182LGY5 B4L338 A0A034WMK6 A0A0P6HZV7 B4NDQ5 Q16GH1 V9L4J1 A0A182N1H6 Q29GM1 A0A195C380 B4H0T1 A0A0M3QZG9 A0A1A9XXP9 A0A3R7PJ90 A0A1L8E4C7 A0A1W4ZNP3 F6NX26 A0A2I4CCC3 A0A1B6HJK7 Q3ZLC7 Q802F5 A0A2D0T2A6 A0A2K5IB97 A0A131XPR8 K4G7U2 V9LHW2 A0A2K5KRG9 A0A2R5LBD8 A0A2K5XJK8 G7PUM5
A0A0K8TNU5 A0A182W4P6 A0A2P8XN98 A0A158NDB9 A0A2M4B064 A0A182M1C4 A0A182FK20 A0A2M3ZDR0 A0A2M3ZKG9 A0A084VVL3 W5JL32 A0A2M4C4H7 A0A2M4AZK6 A0A182K4H6 A0A3L8DRU5 A0A182P7X3 A0A1I8P4F5 A0A182XLS0 A0A182IA79 A0A182V3Z8 A0A0L0C241 A0A182Y7I8 A0A1Q3F8E7 A0A023ED07 A0A2M4AZL6 A0A182G7C5 A0A1S4FUI2 Q16P89 A0A1J1HHR8 A0A1I8MC84 B0WSX0 A0A034WNF4 A0A023EEN1 A7L483 A0A023ED31 A0A232ELY3 A0A1B6CDG1 K7J4W5 A0A151WES7 A0A1L8E4I4 B3MZN5 B4PYH3 A0A1L8E460 A0A182TZF6 B4I683 A0A1W4VT44 B4M3C7 U4UUM6 Q9VRA0 B3NWJ7 D6WBT4 A0A195FI18 B7FNM1 B4JXP9 A0A0K8UMT8 Q7QFL8 A0A182LGY5 B4L338 A0A034WMK6 A0A0P6HZV7 B4NDQ5 Q16GH1 V9L4J1 A0A182N1H6 Q29GM1 A0A195C380 B4H0T1 A0A0M3QZG9 A0A1A9XXP9 A0A3R7PJ90 A0A1L8E4C7 A0A1W4ZNP3 F6NX26 A0A2I4CCC3 A0A1B6HJK7 Q3ZLC7 Q802F5 A0A2D0T2A6 A0A2K5IB97 A0A131XPR8 K4G7U2 V9LHW2 A0A2K5KRG9 A0A2R5LBD8 A0A2K5XJK8 G7PUM5
PDB
2LJK
E-value=2.98531e-06,
Score=115
Ontologies
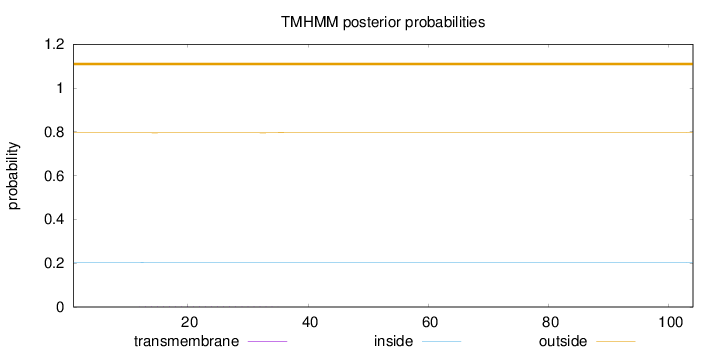
Topology
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03534
Exp number, first 60 AAs:
0.03534
Total prob of N-in:
0.20438
outside
1 - 104
Population Genetic Test Statistics
Pi
218.305485
Theta
169.087073
Tajima's D
0.909109
CLR
1.06775
CSRT
0.634568271586421
Interpretation
Uncertain
