Pre Gene Modal
BGIBMGA012462
Annotation
Molybdenum_cofactor_sulfurase_[Papilio_machaon]
Full name
Molybdenum cofactor sulfurase
+ More
Molybdenum cofactor sulfurase 2
Molybdenum cofactor sulfurase 2
Alternative Name
Molybdenum cofactor sulfurtransferase
Protein maroon-like
Molybdenum cofactor sulfurtransferase 2
Protein maroon-like 2
Protein maroon-like
Molybdenum cofactor sulfurtransferase 2
Protein maroon-like 2
Location in the cell
Nuclear Reliability : 2.953
Sequence
CDS
ATGACCGTGCTAAGTCAAATAATAAAGCCCGATGATATGATAAAAATTACATCAGAGTTTGCAAGACTGGGAGACAGATGTTATTTGGACAATGCCGGTGCTACTTTATACCCAAAAAGTTTGATTACGTCCATAAATGAAGACTTATTAAAAAATGTTTATATGAATCCTCATACAGATAAAAATACAAAAGATTACATAGAACAAATTAGATGTCTCATACTAAAACATTTCAATACAGATCCTTCAACATATACGTTGATTTTCACATCTGGAACCACTCAAGCCTTGAAATTAGTTATTGAATCATTTCAATTTATGAAGAATGAAGATGATGATTTAAATTGTGGTTCCTTTGTTTACCTAGAAGATAATCACACATCTGTTGTTGGCTTAAGAGAACTTGCAGTAGACAAAGATGCAGAAGTAGTCCACATAGCCCATGAAGATTTCTTAAATGTCATAAACACGAAAGCCAAACAAACAAGCAAATACACAAATGGTGGTAACAGTCTTGTTGCTTATCCTGCTCAGAGTAATTTCAATGGTTTTAAATACCCTTTAAACTGCATAGAGAATATAAAAAATGGATGTCTGAACAATCACTTGAAAAAGCATTTATGTGAGATAAACAGTGACTGGTACGTTCTGTTAGATGCTGCTGCATATGTTGCTACCAGTAAATTGGATCTCACAAAAGTGCAACCAGATTTTGTATCTTTATCGTTTTATAAGATTTTTGGTTTCCCAACTGGATTGGGAGCTTTATTAGTTAAAAAATCTAGTGAGAATGTTTTAAGCCAAAAAAGGTATTTTGGTGGAGGCACTGTTGATGCATTATTGAGTAATGAGCACTATCATATCAAAAGGGAAATCTTTCATGAAAGATTTGAAGATGGATCTCTATCTTTCTTATCCATAATTTCTTTAAAACAATGTCTGGATACAATGTACAGAATCATACCAAGAATAATACATGACGATATTATGGAAACAATATCTTACCATACGTTTTATTTAGCAAAAGATTTGTATTGTCAGCTATTAGATTTACGACATCGAAACGGCACAAAGGCTATCAAATTCTATTTAGATTCAGATTTTAGTGACATTACAAAGCAAGGGGGCGTTTTAACTTTTAATCTGGTGAGAGAAGATGGAACTTACATTGGTTTTTCTGAGTTCCAACACATGGCAGACTTATTTAACATAAGTGTCAGAACAGGATGTTTTTGTAATTCTGGATCATGTCAAAGACATTTGCACATGTCAAACAAGGATATGAAAGATATGTATAATGCTGGTCACAGATGTGGAGATGAAGTAGATTTAATTAATGGAAAGCCCACAGGTTGGAACATAGGTCCAAAAGGTTTTGAATACGACCGAGAATGGATGATAGTTAAAGACAATGGAGTGTGTCTCACACAGAAACAGAATACACGTATGTGCATGATTCGCCCACAAATCGATCTCAAACTAAAAGTAATGATATTAAATTTCCCAGGTAAAACTCCCATATCCATACCCTTGGAAAATTCTATAAACGAGGTACAGAAAAATGGATCACTCTGTCATAGTAAAGTCTGTACAGATATGATTAAGGGCATTGATTGTGGTGATGAAGTTGCAGACTGGATATCTGAAGCTCTTGAAGTGTCATTCCTTCGTTTAATACGACAATCTAGCAACGACAACAGATCTCTAAAGAAGAAGAAAGATGAAGATAAAAAATTATTGTCGCTTAGCAATCAGGCTCAATATCTTCTAATTAATAAAGCAACAGTTAAATGGCTTAGTGAAAAGATCAAAGATCCTCTCTTTACTGATGATTTAAATCATTTAACAGACCGATTTAGAGGGAATCTCATTATAGAAATGGAACAAGAATTGTTGGAACGTGAATGGCACAGTGTTATAATTGGAAATCACGAGTTTAAGGTGGAAGGGCAGTGCCCCAGGTGTCAAATGGTATGCATAGATCAACAAACTGGCGAGAAAACTGTAGAGCCCCTTAGGACAATAGCAGAACAATTTGGAGGGAAACTTCGATTTGGTATTTACTTGAGTTATGTTGGCACAGTGAATAAATCTGACGACAGAACTTTGAAAACTTATTCTCCAATTAAAGCAATACTAAATGATGACAACATCAGTAGATGA
Protein
MTVLSQIIKPDDMIKITSEFARLGDRCYLDNAGATLYPKSLITSINEDLLKNVYMNPHTDKNTKDYIEQIRCLILKHFNTDPSTYTLIFTSGTTQALKLVIESFQFMKNEDDDLNCGSFVYLEDNHTSVVGLRELAVDKDAEVVHIAHEDFLNVINTKAKQTSKYTNGGNSLVAYPAQSNFNGFKYPLNCIENIKNGCLNNHLKKHLCEINSDWYVLLDAAAYVATSKLDLTKVQPDFVSLSFYKIFGFPTGLGALLVKKSSENVLSQKRYFGGGTVDALLSNEHYHIKREIFHERFEDGSLSFLSIISLKQCLDTMYRIIPRIIHDDIMETISYHTFYLAKDLYCQLLDLRHRNGTKAIKFYLDSDFSDITKQGGVLTFNLVREDGTYIGFSEFQHMADLFNISVRTGCFCNSGSCQRHLHMSNKDMKDMYNAGHRCGDEVDLINGKPTGWNIGPKGFEYDREWMIVKDNGVCLTQKQNTRMCMIRPQIDLKLKVMILNFPGKTPISIPLENSINEVQKNGSLCHSKVCTDMIKGIDCGDEVADWISEALEVSFLRLIRQSSNDNRSLKKKKDEDKKLLSLSNQAQYLLINKATVKWLSEKIKDPLFTDDLNHLTDRFRGNLIIEMEQELLEREWHSVIIGNHEFKVEGQCPRCQMVCIDQQTGEKTVEPLRTIAEQFGGKLRFGIYLSYVGTVNKSDDRTLKTYSPIKAILNDDNISR
Summary
Description
Sulfurates the molybdenum cofactor. Sulfation of molybdenum is essential for xanthine dehydrogenase (XDH) and aldehyde oxidase (ADO) enzymes in which molybdenum cofactor is liganded by 1 oxygen and 1 sulfur atom in active form.
Catalytic Activity
AH2 + L-cysteine + Mo-molybdopterin = A + H2O + L-alanine + thio-Mo-molybdopterin
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family. MOCOS subfamily.
Keywords
Complete proteome
Molybdenum cofactor biosynthesis
Pyridoxal phosphate
Reference proteome
Transferase
Phosphoprotein
Feature
chain Molybdenum cofactor sulfurase 2
Uniprot
EC Number
2.8.1.9
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
5VPR
E-value=4.55525e-11,
Score=166
Ontologies
GO
Topology
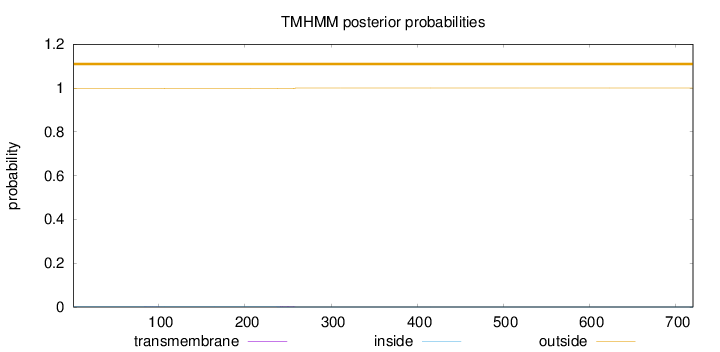
Length:
720
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0369100000000001
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00169
outside
1 - 720
Population Genetic Test Statistics
Pi
243.725271
Theta
221.265932
Tajima's D
0.288613
CLR
0.001795
CSRT
0.455027248637568
Interpretation
Uncertain
