Pre Gene Modal
BGIBMGA012503
Annotation
PREDICTED:_poly(rC)-binding_protein_3_isoform_X3_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.092 Nuclear Reliability : 1.633
Sequence
CDS
ATGGACCTGGAGAAAGCCCCTTCTCTCCATGAGGAAGACTCAAAAGTTATCCTCACTATCAGACTGATTATGCAGGGGAAGGAAGTTGGGAGCATTATTGGAAAAAAAGGAGAGATTGTTAAAAGATTTAGAGAAGAGTCCGGTGCTAAAATTAATATTTCTGATGGATCATGCCCGGAGCGTATTGTTACTGTCACAGGAAACACAACTGCTATATTCAAGGCATTCACACTCATATGCAAAAAATTTGAAGAGTGGTGCACTCAGTTCAATGAGGGCGGAGGCGGCGGATCGCGTGCTCCCATCACGCTGCGGCTTATTGTGCCCGCCTCGCAGTGTGGCTCACTTATTGGAAAGGGGGGCTCAAAAATCAAGGAAATCCGTGATGTCACTGGTGCCAACATTCAGGTGGCGAGCGAAATGTTGCCGAATTCAACTGAACGAGCGGTGACAATCAGTGGAACCTGTGACGCTATAACCCAATGCATTTATCATATTTGTTGTGTTATGTTAGAGAGCCCTCCGAAGGGAGCAACAATCCCGTACAGACCAAAGCCAAATGTAGCGGGACCTGTGATTCTAGCCGGCGGACAAGCCTACACTATCCAAGGAAACTATGCCGTCCCAGCCCAAGACATGGGAGGGCTCGCTAAGTCGCCGTTGGCCAGTCTCGCTGCCCTGGGCCTCGGTGGTCTCGGACCTGGGAATGCTGCAGGATTAAATCCTGCTGCACTGGCGGCTCTGGCCGGCTCGCAGCTGCGCACTACGAACCAGTCCCGCAACCAGCCCGCCACCAACCAGCAGTCGCACGAGATGACCGTGCCCAATGAGCTCATCGGGTGCATCATCGGAAAGGGCGGCACCAAGATCGCTGAAATCCGTCAAATATCCGGCGCAATGATCAGGATATCCAACTGCGAGGAGCGCGAGGGCGGCAGCACCGACCGGACCATCACTATAACTGGCAACCCCGACTCGGTGGCGTTGGCGCAGTACCTCATCAACATGAGGATCACGATGGAGACGGCCGGCCTCCCGCTGCCCGGCTACCACTACATCGCTCCGAATCACATAGTGAAAGCGCCGATACATTGA
Protein
MDLEKAPSLHEEDSKVILTIRLIMQGKEVGSIIGKKGEIVKRFREESGAKINISDGSCPERIVTVTGNTTAIFKAFTLICKKFEEWCTQFNEGGGGGSRAPITLRLIVPASQCGSLIGKGGSKIKEIRDVTGANIQVASEMLPNSTERAVTISGTCDAITQCIYHICCVMLESPPKGATIPYRPKPNVAGPVILAGGQAYTIQGNYAVPAQDMGGLAKSPLASLAALGLGGLGPGNAAGLNPAALAALAGSQLRTTNQSRNQPATNQQSHEMTVPNELIGCIIGKGGTKIAEIRQISGAMIRISNCEEREGGSTDRTITITGNPDSVALAQYLINMRITMETAGLPLPGYHYIAPNHIVKAPIH
Summary
Uniprot
A0A3S2M6Y9
A0A1Y1L8J0
V5GMR9
S4P396
A0A2J7QDK8
A0A1Y1LEN0
+ More
A0A0C9S1H0 A0A0C9QZE8 T1E2R2 A0A1W4XG91 T1E2R8 K7IYD3 A0A2M4AQ93 A0A2M4CTT4 A0A1L8DGU8 A0A2A3E5V3 T1PMM0 A0A1B6LFG9 A0A0A1XHH2 A0A0A1WFP7 A0A0K8TVT8 A0A034W8N0 A0A1I8NW24 Q5TRZ9 A0A084WCZ9 A0A336MH90 B4N4G9 B4LC01 B4L003 A0A2M4AQB1 B4J0R1 A0A336MBM9 A0A1W4VAV9 A0A0J9URC1 B4PI46 B3NJ10 Q95SZ9 A0A3B0JRT1 A0A1W4UYL4 A0A0J9RZT7 M9PIK1 A0A0R1E3W1 A0A0Q5UK39 A0A1Y1L8V2 A0A310SSG3 D6WBV2 A0A2A3E5Y9 V5G5P0 A0A0T6BF49 A0A1I8NW47 A0A2J7QDL3 A0A0A1XLI1 A0A034W8M5 A0A1J1I786 E2BX15 A0A088ATQ8 A0A2J7QDL4 A0A0Q9X2D2 A0A0Q9WIZ9 A0A0Q9XQU7 A0A1Y1LAY0 A0A1W4VBB5 A0A0J9S140 A0A0R1DZI6 A0A0Q5UHQ7 A4V2A4 U5ESU4 A0A0C9R4I1 A0A146M848 A0A1Q3EU26 A0A1B6KVV4 A0A023F506 A0A069DUG0 A0A0P4VKU7 T1HGV2 A0A224XJW6 A0A0V0G963 A0A2M4CL72 A0A182FLA4 A0A0K8W3L8 A0A1I8MHP4 A0A1L8DH25 A0A1I8NW21 A0A1Q3FTK0 A0A034W5Y2 A0A034W8P1 E9G1N5 A0A3B0J860 A0A162T5T0 A0A336MFK0 A0A1W4VC80 A0A0J9S150 A0A1W4VAV4 A0A3B0JUK4 A0A0Q5U684 Q0E8A7 A0A0R1DZH1 A0A1W4UYZ8
A0A0C9S1H0 A0A0C9QZE8 T1E2R2 A0A1W4XG91 T1E2R8 K7IYD3 A0A2M4AQ93 A0A2M4CTT4 A0A1L8DGU8 A0A2A3E5V3 T1PMM0 A0A1B6LFG9 A0A0A1XHH2 A0A0A1WFP7 A0A0K8TVT8 A0A034W8N0 A0A1I8NW24 Q5TRZ9 A0A084WCZ9 A0A336MH90 B4N4G9 B4LC01 B4L003 A0A2M4AQB1 B4J0R1 A0A336MBM9 A0A1W4VAV9 A0A0J9URC1 B4PI46 B3NJ10 Q95SZ9 A0A3B0JRT1 A0A1W4UYL4 A0A0J9RZT7 M9PIK1 A0A0R1E3W1 A0A0Q5UK39 A0A1Y1L8V2 A0A310SSG3 D6WBV2 A0A2A3E5Y9 V5G5P0 A0A0T6BF49 A0A1I8NW47 A0A2J7QDL3 A0A0A1XLI1 A0A034W8M5 A0A1J1I786 E2BX15 A0A088ATQ8 A0A2J7QDL4 A0A0Q9X2D2 A0A0Q9WIZ9 A0A0Q9XQU7 A0A1Y1LAY0 A0A1W4VBB5 A0A0J9S140 A0A0R1DZI6 A0A0Q5UHQ7 A4V2A4 U5ESU4 A0A0C9R4I1 A0A146M848 A0A1Q3EU26 A0A1B6KVV4 A0A023F506 A0A069DUG0 A0A0P4VKU7 T1HGV2 A0A224XJW6 A0A0V0G963 A0A2M4CL72 A0A182FLA4 A0A0K8W3L8 A0A1I8MHP4 A0A1L8DH25 A0A1I8NW21 A0A1Q3FTK0 A0A034W5Y2 A0A034W8P1 E9G1N5 A0A3B0J860 A0A162T5T0 A0A336MFK0 A0A1W4VC80 A0A0J9S150 A0A1W4VAV4 A0A3B0JUK4 A0A0Q5U684 Q0E8A7 A0A0R1DZH1 A0A1W4UYZ8
Pubmed
EMBL
RSAL01000018
RVE52888.1
GEZM01062144
GEZM01062140
JAV70013.1
GALX01003072
+ More
JAB65394.1 GAIX01006534 JAA86026.1 NEVH01015820 PNF26668.1 GEZM01062142 GEZM01062141 GEZM01062139 JAV70016.1 GBYB01014621 JAG84388.1 GBYB01009169 GBYB01014623 JAG78936.1 JAG84390.1 GALA01001065 JAA93787.1 GALA01001050 JAA93802.1 GGFK01009630 MBW42951.1 GGFL01004555 MBW68733.1 GFDF01008401 JAV05683.1 KZ288357 PBC27117.1 KA649345 AFP63974.1 GEBQ01017549 JAT22428.1 GBXI01003885 JAD10407.1 GBXI01016570 JAC97721.1 GDHF01033933 JAI18381.1 GAKP01008836 GAKP01008832 GAKP01008824 JAC50128.1 AAAB01008948 EAL40201.3 ATLV01022904 ATLV01022905 KE525337 KFB48093.1 UFQS01000865 UFQT01000865 SSX07355.1 SSX27697.1 CH964095 EDW79043.2 CH940647 EDW69801.2 CH933809 EDW19038.1 GGFK01009632 MBW42953.1 CH916366 EDV97916.1 SSX07353.1 SSX27695.1 CM002912 KMZ01131.1 CM000159 EDW95494.1 CH954178 EDV52727.1 AY060403 AE014296 AAL25442.1 AAS65095.1 OUUW01000002 SPP76409.1 KMZ01129.1 AGB94891.1 KRK02465.1 KQS44406.1 GEZM01062143 GEZM01062138 JAV70014.1 KQ760559 OAD59806.1 KQ971314 EEZ97865.2 PBC27118.1 GALX01003071 JAB65395.1 LJIG01000976 KRT85964.1 PNF26671.1 GBXI01002859 JAD11433.1 GAKP01008829 GAKP01008826 JAC50123.1 CVRI01000043 CRK96035.1 GL451202 EFN79734.1 PNF26667.1 KRF98994.1 KRF84584.1 KRG06472.1 GEZM01062137 JAV70021.1 KMZ01120.1 KRK02464.1 KQS44403.1 AAN12194.2 AGB94890.1 GANO01003071 JAB56800.1 GBYB01011204 JAG80971.1 GDHC01002685 JAQ15944.1 GFDL01016231 JAV18814.1 GEBQ01024391 JAT15586.1 GBBI01002603 JAC16109.1 GBGD01001458 JAC87431.1 GDKW01001621 JAI54974.1 ACPB03025788 GFTR01006308 JAW10118.1 GECL01001582 JAP04542.1 GGFL01001934 MBW66112.1 GDHF01006556 JAI45758.1 GFDF01008331 JAV05753.1 GFDL01004121 JAV30924.1 GAKP01008828 JAC50124.1 GAKP01008830 GAKP01008814 JAC50138.1 GL732529 EFX86525.1 SPP76413.1 LRGB01000007 KZS21933.1 SSX07357.1 SSX27699.1 KMZ01130.1 SPP76411.1 KQS44411.1 ABI31266.1 KRK02459.1
JAB65394.1 GAIX01006534 JAA86026.1 NEVH01015820 PNF26668.1 GEZM01062142 GEZM01062141 GEZM01062139 JAV70016.1 GBYB01014621 JAG84388.1 GBYB01009169 GBYB01014623 JAG78936.1 JAG84390.1 GALA01001065 JAA93787.1 GALA01001050 JAA93802.1 GGFK01009630 MBW42951.1 GGFL01004555 MBW68733.1 GFDF01008401 JAV05683.1 KZ288357 PBC27117.1 KA649345 AFP63974.1 GEBQ01017549 JAT22428.1 GBXI01003885 JAD10407.1 GBXI01016570 JAC97721.1 GDHF01033933 JAI18381.1 GAKP01008836 GAKP01008832 GAKP01008824 JAC50128.1 AAAB01008948 EAL40201.3 ATLV01022904 ATLV01022905 KE525337 KFB48093.1 UFQS01000865 UFQT01000865 SSX07355.1 SSX27697.1 CH964095 EDW79043.2 CH940647 EDW69801.2 CH933809 EDW19038.1 GGFK01009632 MBW42953.1 CH916366 EDV97916.1 SSX07353.1 SSX27695.1 CM002912 KMZ01131.1 CM000159 EDW95494.1 CH954178 EDV52727.1 AY060403 AE014296 AAL25442.1 AAS65095.1 OUUW01000002 SPP76409.1 KMZ01129.1 AGB94891.1 KRK02465.1 KQS44406.1 GEZM01062143 GEZM01062138 JAV70014.1 KQ760559 OAD59806.1 KQ971314 EEZ97865.2 PBC27118.1 GALX01003071 JAB65395.1 LJIG01000976 KRT85964.1 PNF26671.1 GBXI01002859 JAD11433.1 GAKP01008829 GAKP01008826 JAC50123.1 CVRI01000043 CRK96035.1 GL451202 EFN79734.1 PNF26667.1 KRF98994.1 KRF84584.1 KRG06472.1 GEZM01062137 JAV70021.1 KMZ01120.1 KRK02464.1 KQS44403.1 AAN12194.2 AGB94890.1 GANO01003071 JAB56800.1 GBYB01011204 JAG80971.1 GDHC01002685 JAQ15944.1 GFDL01016231 JAV18814.1 GEBQ01024391 JAT15586.1 GBBI01002603 JAC16109.1 GBGD01001458 JAC87431.1 GDKW01001621 JAI54974.1 ACPB03025788 GFTR01006308 JAW10118.1 GECL01001582 JAP04542.1 GGFL01001934 MBW66112.1 GDHF01006556 JAI45758.1 GFDF01008331 JAV05753.1 GFDL01004121 JAV30924.1 GAKP01008828 JAC50124.1 GAKP01008830 GAKP01008814 JAC50138.1 GL732529 EFX86525.1 SPP76413.1 LRGB01000007 KZS21933.1 SSX07357.1 SSX27699.1 KMZ01130.1 SPP76411.1 KQS44411.1 ABI31266.1 KRK02459.1
Proteomes
UP000283053
UP000235965
UP000192223
UP000002358
UP000242457
UP000095301
+ More
UP000095300 UP000007062 UP000030765 UP000007798 UP000008792 UP000009192 UP000001070 UP000192221 UP000002282 UP000008711 UP000000803 UP000268350 UP000007266 UP000183832 UP000008237 UP000005203 UP000015103 UP000069272 UP000000305 UP000076858
UP000095300 UP000007062 UP000030765 UP000007798 UP000008792 UP000009192 UP000001070 UP000192221 UP000002282 UP000008711 UP000000803 UP000268350 UP000007266 UP000183832 UP000008237 UP000005203 UP000015103 UP000069272 UP000000305 UP000076858
Pfam
PF00013 KH_1
SUPFAM
SSF54791
SSF54791
Gene 3D
ProteinModelPortal
A0A3S2M6Y9
A0A1Y1L8J0
V5GMR9
S4P396
A0A2J7QDK8
A0A1Y1LEN0
+ More
A0A0C9S1H0 A0A0C9QZE8 T1E2R2 A0A1W4XG91 T1E2R8 K7IYD3 A0A2M4AQ93 A0A2M4CTT4 A0A1L8DGU8 A0A2A3E5V3 T1PMM0 A0A1B6LFG9 A0A0A1XHH2 A0A0A1WFP7 A0A0K8TVT8 A0A034W8N0 A0A1I8NW24 Q5TRZ9 A0A084WCZ9 A0A336MH90 B4N4G9 B4LC01 B4L003 A0A2M4AQB1 B4J0R1 A0A336MBM9 A0A1W4VAV9 A0A0J9URC1 B4PI46 B3NJ10 Q95SZ9 A0A3B0JRT1 A0A1W4UYL4 A0A0J9RZT7 M9PIK1 A0A0R1E3W1 A0A0Q5UK39 A0A1Y1L8V2 A0A310SSG3 D6WBV2 A0A2A3E5Y9 V5G5P0 A0A0T6BF49 A0A1I8NW47 A0A2J7QDL3 A0A0A1XLI1 A0A034W8M5 A0A1J1I786 E2BX15 A0A088ATQ8 A0A2J7QDL4 A0A0Q9X2D2 A0A0Q9WIZ9 A0A0Q9XQU7 A0A1Y1LAY0 A0A1W4VBB5 A0A0J9S140 A0A0R1DZI6 A0A0Q5UHQ7 A4V2A4 U5ESU4 A0A0C9R4I1 A0A146M848 A0A1Q3EU26 A0A1B6KVV4 A0A023F506 A0A069DUG0 A0A0P4VKU7 T1HGV2 A0A224XJW6 A0A0V0G963 A0A2M4CL72 A0A182FLA4 A0A0K8W3L8 A0A1I8MHP4 A0A1L8DH25 A0A1I8NW21 A0A1Q3FTK0 A0A034W5Y2 A0A034W8P1 E9G1N5 A0A3B0J860 A0A162T5T0 A0A336MFK0 A0A1W4VC80 A0A0J9S150 A0A1W4VAV4 A0A3B0JUK4 A0A0Q5U684 Q0E8A7 A0A0R1DZH1 A0A1W4UYZ8
A0A0C9S1H0 A0A0C9QZE8 T1E2R2 A0A1W4XG91 T1E2R8 K7IYD3 A0A2M4AQ93 A0A2M4CTT4 A0A1L8DGU8 A0A2A3E5V3 T1PMM0 A0A1B6LFG9 A0A0A1XHH2 A0A0A1WFP7 A0A0K8TVT8 A0A034W8N0 A0A1I8NW24 Q5TRZ9 A0A084WCZ9 A0A336MH90 B4N4G9 B4LC01 B4L003 A0A2M4AQB1 B4J0R1 A0A336MBM9 A0A1W4VAV9 A0A0J9URC1 B4PI46 B3NJ10 Q95SZ9 A0A3B0JRT1 A0A1W4UYL4 A0A0J9RZT7 M9PIK1 A0A0R1E3W1 A0A0Q5UK39 A0A1Y1L8V2 A0A310SSG3 D6WBV2 A0A2A3E5Y9 V5G5P0 A0A0T6BF49 A0A1I8NW47 A0A2J7QDL3 A0A0A1XLI1 A0A034W8M5 A0A1J1I786 E2BX15 A0A088ATQ8 A0A2J7QDL4 A0A0Q9X2D2 A0A0Q9WIZ9 A0A0Q9XQU7 A0A1Y1LAY0 A0A1W4VBB5 A0A0J9S140 A0A0R1DZI6 A0A0Q5UHQ7 A4V2A4 U5ESU4 A0A0C9R4I1 A0A146M848 A0A1Q3EU26 A0A1B6KVV4 A0A023F506 A0A069DUG0 A0A0P4VKU7 T1HGV2 A0A224XJW6 A0A0V0G963 A0A2M4CL72 A0A182FLA4 A0A0K8W3L8 A0A1I8MHP4 A0A1L8DH25 A0A1I8NW21 A0A1Q3FTK0 A0A034W5Y2 A0A034W8P1 E9G1N5 A0A3B0J860 A0A162T5T0 A0A336MFK0 A0A1W4VC80 A0A0J9S150 A0A1W4VAV4 A0A3B0JUK4 A0A0Q5U684 Q0E8A7 A0A0R1DZH1 A0A1W4UYZ8
PDB
2JZX
E-value=3.55117e-44,
Score=448
Ontologies
GO
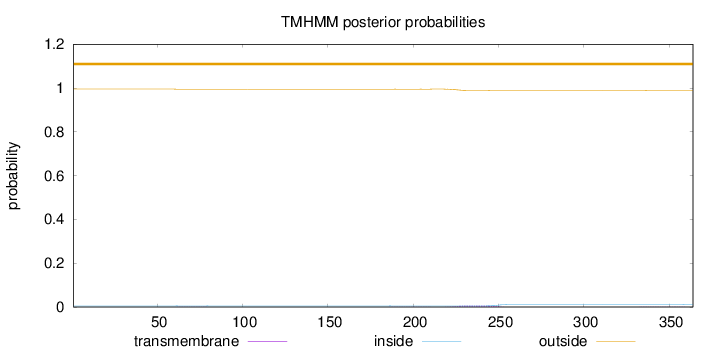
Topology
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18313
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.00493
outside
1 - 364
Population Genetic Test Statistics
Pi
241.077351
Theta
176.284668
Tajima's D
1.249709
CLR
0.035881
CSRT
0.725663716814159
Interpretation
Uncertain
