Gene
KWMTBOMO05114
Annotation
PREDICTED:_glutamate-gated_chloride_channel_isoform_X17_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.283
Sequence
CDS
ATGGGTTGGTCATGCGTCGTGGCACGCGCCGTGGCCTTTATCCTCATGCTCGGCAAAATATCAGCTTTTACGTCCGACATCTTTGCCGCGGGAAAGTCGGACAAAGAGATTTTGGACAATCTTCTTAAAAATACCCGCTACGACAAAAGACTGCTCCCACCAGTTGATGATCCAGAATTCTGTTGTGGTCTAACTTCACCCAACGACTCTTTGGCACAAAATAGGGTCGGGTTTTCGTCGCTTCGGCCCCGTTCGCACAATCGCGGTGTTCTCACAGTAAATGTTAGCGTGCTTCTCCTTAGCTTAGCATCTCCAGATGAATCTAGTCTTAAATACGAAGTGGAGTTTTTACTTCAACAACAATGGTACGACCCTCGGTTAAGATACTCCAACCAATCGCATTACGATTATTTAAATGCCATACATCATCATGAAGATATTTGGCTGCCTGATACATATTTCATCATGCATGGAGATTTTAAGGATCCCATAATACCGATGCATTTTGCGTTACGTATTTATCGCAATGGTACAATTAATTATTTGATGAGGCGACATCTCATACTGTCCTGTCAGGGTCGGCTTAATATATTTCCATTCGATGACCCATTGTGTTCATTTGCCTTAGAAAGTATCTCGTATGAACAATCTGCCATAACATACGTCTGGAAGAATGATGAAGATACACTTAGAAAATCACCATCACTGACGACACTGAACGCCTACCTCATTCAGAACCAGACTATACCCTGTCCAATAAAAGCTAGTTGGAGAGGTAACTACAGCTGTCTAAAGGTTGACTTAATCTTTACTAGAGACCGAGCGTTCTACTTTACTACAGTATTTATTCCTGGAATAATTCTGGTGACATCTTCGTTCATCACATTTTGGCTGGAATGGAACGCGGTGCCAGCCCGTTCCATGATAGGTGTAACAACAATGTTGAACTTTTTTACAACATCTAACGGGTTCCGATCAACTTTGCCAGTAGTATCGAACCTAACAGCCATGAATGTTTGGGACGGTGTATGCATGTGTTTCATTTACGCTTCGCTACTTGAGTTCGTGTGCGTGAACTACGTCGGGAGGAAGAGACCACTACACAATGTGGTATACAGACCAGGGGAGAATCCTGTGACACAGCGACTGCCCGCAGTCCTGAGCAGAATTGGCATTATACTGGCCAGCCCCTTGGAGGCGATGGCATTTCTTCAATGGGTCAAATCTGAGACCCCTCAACCTGCGTGTAGCGGCGAAGGTGATAAGAAGCGAGAATCAACAGGAGCAGCAGATTTGGTGTCGTGTACTGCGTGTACCGGCCCCCCCGGCTCCTGTACGCACACCGCCAATAATGGCGGTGTATCCGAGCCATGTTTCGTCCAGGTTCGCAAAAAAGAACCCCCGCATCCGATCCGAGTGGCGAAGACTATTGACGTAATCGCCAGGATTACGTTTCCAACTGCGTACGCTGTCTTTCTCATCTTCTTCTTCATTCACTACAAAGCGTTTTCTTAA
Protein
MGWSCVVARAVAFILMLGKISAFTSDIFAAGKSDKEILDNLLKNTRYDKRLLPPVDDPEFCCGLTSPNDSLAQNRVGFSSLRPRSHNRGVLTVNVSVLLLSLASPDESSLKYEVEFLLQQQWYDPRLRYSNQSHYDYLNAIHHHEDIWLPDTYFIMHGDFKDPIIPMHFALRIYRNGTINYLMRRHLILSCQGRLNIFPFDDPLCSFALESISYEQSAITYVWKNDEDTLRKSPSLTTLNAYLIQNQTIPCPIKASWRGNYSCLKVDLIFTRDRAFYFTTVFIPGIILVTSSFITFWLEWNAVPARSMIGVTTMLNFFTTSNGFRSTLPVVSNLTAMNVWDGVCMCFIYASLLEFVCVNYVGRKRPLHNVVYRPGENPVTQRLPAVLSRIGIILASPLEAMAFLQWVKSETPQPACSGEGDKKRESTGAADLVSCTACTGPPGSCTHTANNGGVSEPCFVQVRKKEPPHPIRVAKTIDVIARITFPTAYAVFLIFFFIHYKAFS
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A0A1Y1CBU9
A0A1Y1CBW9
A0A194PEA8
A0A1Y1CCN0
A0A1Y1CCI3
A0A1Y1CBZ2
+ More
A0A1Y1CBV1 A0A1Y1CBS9 A0A1Y1CBY9 A0A1Y1CC00 A0A1Y1CBU2 A0A1Y1CBX8 A0A1Y1CCP0 A0A1Y1CBZ5 A0A1Y1CBW7 A0A1Y1CEU1 A0A1Y1CBW1 A0A1Y1CBY5 A0A1Y1CBV9 A0A2A4J638 A0A1Y1CEV2 A0A1Y1CBT8 A0A1Y1CBV2 A0A1Y1CBV3 A0A1Y1CC06 A0A1Y1CBU6 A0A1Y1CCH5 A0A1Y1CBW2 A0A1Y1CC11 A0A1Y1CBY4 A0A1Y1CBW0 A0A1Y1CBT6 A0A336LYA0 A0A1Y1CBV7 A0A1S4GPQ0 Q7Q6Y7 A0A1Y1CEW2 A0A336M5E5 A0A1S4GPR9 A0A1S4GQD2 A0A1S4GQH6 A7UTS4 A0A336LTH9 A0A1W4VAJ8 A0A2A4J4R8 A0A0R1DZ29 A0A0P8XUJ1 A0A0Q9WL63 A0A0J9RVX2 Q2PDY9 A0A0Q5U8V3 A0A0P8ZUZ0 A0A0J9RX89 A0A0R3P3J7 M9PCN0 A0A0J9RVV6 A0A0Q9WUB4 A0A0P8YHI4 A0A0Q5UH41 A0A0J9RVW2 A0A1S4GPQ4 Q5D6W1 A0A0R1DYK3 A0A1I8NFQ3 A0A1I8NFR6 A0A1I8NFP9 A0A1I8NFS0 A0A1Y1N3S0 A0A1W4WUS7 X2JCG3 A0A1W4VNA9 A0A0R1DYI1 M9PFD4 A0A0R1DYH9 A0A0J9RW86 Q2PDZ0 A0A0J9RX61 A0A023F372 Q0GQQ8 A0A0N8P0Y2 A0A0Q9WKP9 A0A0Q5UJG1 M9NFY2 A0A0J9RX66 A0A0R1E3B3 A0A0Q5U541 A0A0J9RW88 A0A0R3P7H5 M9PFD7 A0A0Q9WKQ1 X2JGQ0 A0A0Q5UJE3 A0A0P9AKL0 A0A0J9RW83 A0A0P9AKK1 A0A0Q5U5A6 A0A0J9RVV2 M9NEB0 A0A0J9RVU6 M9PFR7
A0A1Y1CBV1 A0A1Y1CBS9 A0A1Y1CBY9 A0A1Y1CC00 A0A1Y1CBU2 A0A1Y1CBX8 A0A1Y1CCP0 A0A1Y1CBZ5 A0A1Y1CBW7 A0A1Y1CEU1 A0A1Y1CBW1 A0A1Y1CBY5 A0A1Y1CBV9 A0A2A4J638 A0A1Y1CEV2 A0A1Y1CBT8 A0A1Y1CBV2 A0A1Y1CBV3 A0A1Y1CC06 A0A1Y1CBU6 A0A1Y1CCH5 A0A1Y1CBW2 A0A1Y1CC11 A0A1Y1CBY4 A0A1Y1CBW0 A0A1Y1CBT6 A0A336LYA0 A0A1Y1CBV7 A0A1S4GPQ0 Q7Q6Y7 A0A1Y1CEW2 A0A336M5E5 A0A1S4GPR9 A0A1S4GQD2 A0A1S4GQH6 A7UTS4 A0A336LTH9 A0A1W4VAJ8 A0A2A4J4R8 A0A0R1DZ29 A0A0P8XUJ1 A0A0Q9WL63 A0A0J9RVX2 Q2PDY9 A0A0Q5U8V3 A0A0P8ZUZ0 A0A0J9RX89 A0A0R3P3J7 M9PCN0 A0A0J9RVV6 A0A0Q9WUB4 A0A0P8YHI4 A0A0Q5UH41 A0A0J9RVW2 A0A1S4GPQ4 Q5D6W1 A0A0R1DYK3 A0A1I8NFQ3 A0A1I8NFR6 A0A1I8NFP9 A0A1I8NFS0 A0A1Y1N3S0 A0A1W4WUS7 X2JCG3 A0A1W4VNA9 A0A0R1DYI1 M9PFD4 A0A0R1DYH9 A0A0J9RW86 Q2PDZ0 A0A0J9RX61 A0A023F372 Q0GQQ8 A0A0N8P0Y2 A0A0Q9WKP9 A0A0Q5UJG1 M9NFY2 A0A0J9RX66 A0A0R1E3B3 A0A0Q5U541 A0A0J9RW88 A0A0R3P7H5 M9PFD7 A0A0Q9WKQ1 X2JGQ0 A0A0Q5UJE3 A0A0P9AKL0 A0A0J9RW83 A0A0P9AKK1 A0A0Q5U5A6 A0A0J9RVV2 M9NEB0 A0A0J9RVU6 M9PFR7
Pubmed
EMBL
LC259042
BAX77806.1
LC259063
BAX77827.1
KQ459606
KPI91363.1
+ More
LC259036 BAX77800.1 LC259047 BAX77811.1 LC259069 BAX77833.1 LC259041 BAX77805.1 LC259032 BAX77796.1 LC259034 BAX77798.1 LC259044 BAX77808.1 LC259031 BAX77795.1 LC259070 BAX77834.1 LC259046 BAX77810.1 LC259048 BAX77812.1 LC259049 BAX77813.1 LC259035 BAX77799.1 LC259051 BAX77815.1 LC259038 BAX77802.1 LC259052 BAX77816.1 NWSH01002991 PCG67138.1 LC259045 BAX77809.1 LC259030 BAX77794.1 LC259029 BAX77793.1 LC259040 BAX77804.1 LC259054 BAX77818.1 LC259043 BAX77807.1 LC259037 BAX77801.1 LC259039 BAX77803.1 LC259068 BAX77832.1 LC259073 BAX77837.1 LC259050 BAX77814.1 LC259033 BAX77797.1 UFQT01000185 SSX21247.1 LC259053 BAX77817.1 AAAB01008960 EAA11458.3 LC259055 BAX77819.1 SSX21248.1 EDO63697.1 SSX21250.1 PCG67137.1 CM000159 KRK02143.1 CH902618 KPU78394.1 CH940647 KRF85250.1 CM002912 KMY99858.1 AE014296 ABC66136.2 CH954178 KQS44132.1 KPU78399.1 KMY99869.1 CH379069 KRT07771.1 AGB94587.1 KMY99843.1 KRF85249.1 KPU78406.1 KQS44147.1 KMY99848.1 AY880248 AAX11175.1 KRK02139.1 GEZM01013533 GEZM01013532 JAV92504.1 AHN58101.1 KRK02124.1 AGB94588.1 KRK02128.1 KMY99852.1 ABC66135.2 KMY99844.1 GBBI01003101 JAC15611.1 DQ667191 ABG75743.1 KPU78396.1 KRF85251.1 KQS44150.1 AFH04453.1 KMY99849.1 KRK02130.1 KQS44152.1 KMY99857.1 KRT07775.1 AGB94593.1 KRF85256.1 AHN58100.1 KQS44134.1 KPU78402.1 KMY99847.1 KPU78393.1 KQS44133.1 KMY99846.1 AFH04452.1 KMY99841.1 AGB94589.1
LC259036 BAX77800.1 LC259047 BAX77811.1 LC259069 BAX77833.1 LC259041 BAX77805.1 LC259032 BAX77796.1 LC259034 BAX77798.1 LC259044 BAX77808.1 LC259031 BAX77795.1 LC259070 BAX77834.1 LC259046 BAX77810.1 LC259048 BAX77812.1 LC259049 BAX77813.1 LC259035 BAX77799.1 LC259051 BAX77815.1 LC259038 BAX77802.1 LC259052 BAX77816.1 NWSH01002991 PCG67138.1 LC259045 BAX77809.1 LC259030 BAX77794.1 LC259029 BAX77793.1 LC259040 BAX77804.1 LC259054 BAX77818.1 LC259043 BAX77807.1 LC259037 BAX77801.1 LC259039 BAX77803.1 LC259068 BAX77832.1 LC259073 BAX77837.1 LC259050 BAX77814.1 LC259033 BAX77797.1 UFQT01000185 SSX21247.1 LC259053 BAX77817.1 AAAB01008960 EAA11458.3 LC259055 BAX77819.1 SSX21248.1 EDO63697.1 SSX21250.1 PCG67137.1 CM000159 KRK02143.1 CH902618 KPU78394.1 CH940647 KRF85250.1 CM002912 KMY99858.1 AE014296 ABC66136.2 CH954178 KQS44132.1 KPU78399.1 KMY99869.1 CH379069 KRT07771.1 AGB94587.1 KMY99843.1 KRF85249.1 KPU78406.1 KQS44147.1 KMY99848.1 AY880248 AAX11175.1 KRK02139.1 GEZM01013533 GEZM01013532 JAV92504.1 AHN58101.1 KRK02124.1 AGB94588.1 KRK02128.1 KMY99852.1 ABC66135.2 KMY99844.1 GBBI01003101 JAC15611.1 DQ667191 ABG75743.1 KPU78396.1 KRF85251.1 KQS44150.1 AFH04453.1 KMY99849.1 KRK02130.1 KQS44152.1 KMY99857.1 KRT07775.1 AGB94593.1 KRF85256.1 AHN58100.1 KQS44134.1 KPU78402.1 KMY99847.1 KPU78393.1 KQS44133.1 KMY99846.1 AFH04452.1 KMY99841.1 AGB94589.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A1Y1CBU9
A0A1Y1CBW9
A0A194PEA8
A0A1Y1CCN0
A0A1Y1CCI3
A0A1Y1CBZ2
+ More
A0A1Y1CBV1 A0A1Y1CBS9 A0A1Y1CBY9 A0A1Y1CC00 A0A1Y1CBU2 A0A1Y1CBX8 A0A1Y1CCP0 A0A1Y1CBZ5 A0A1Y1CBW7 A0A1Y1CEU1 A0A1Y1CBW1 A0A1Y1CBY5 A0A1Y1CBV9 A0A2A4J638 A0A1Y1CEV2 A0A1Y1CBT8 A0A1Y1CBV2 A0A1Y1CBV3 A0A1Y1CC06 A0A1Y1CBU6 A0A1Y1CCH5 A0A1Y1CBW2 A0A1Y1CC11 A0A1Y1CBY4 A0A1Y1CBW0 A0A1Y1CBT6 A0A336LYA0 A0A1Y1CBV7 A0A1S4GPQ0 Q7Q6Y7 A0A1Y1CEW2 A0A336M5E5 A0A1S4GPR9 A0A1S4GQD2 A0A1S4GQH6 A7UTS4 A0A336LTH9 A0A1W4VAJ8 A0A2A4J4R8 A0A0R1DZ29 A0A0P8XUJ1 A0A0Q9WL63 A0A0J9RVX2 Q2PDY9 A0A0Q5U8V3 A0A0P8ZUZ0 A0A0J9RX89 A0A0R3P3J7 M9PCN0 A0A0J9RVV6 A0A0Q9WUB4 A0A0P8YHI4 A0A0Q5UH41 A0A0J9RVW2 A0A1S4GPQ4 Q5D6W1 A0A0R1DYK3 A0A1I8NFQ3 A0A1I8NFR6 A0A1I8NFP9 A0A1I8NFS0 A0A1Y1N3S0 A0A1W4WUS7 X2JCG3 A0A1W4VNA9 A0A0R1DYI1 M9PFD4 A0A0R1DYH9 A0A0J9RW86 Q2PDZ0 A0A0J9RX61 A0A023F372 Q0GQQ8 A0A0N8P0Y2 A0A0Q9WKP9 A0A0Q5UJG1 M9NFY2 A0A0J9RX66 A0A0R1E3B3 A0A0Q5U541 A0A0J9RW88 A0A0R3P7H5 M9PFD7 A0A0Q9WKQ1 X2JGQ0 A0A0Q5UJE3 A0A0P9AKL0 A0A0J9RW83 A0A0P9AKK1 A0A0Q5U5A6 A0A0J9RVV2 M9NEB0 A0A0J9RVU6 M9PFR7
A0A1Y1CBV1 A0A1Y1CBS9 A0A1Y1CBY9 A0A1Y1CC00 A0A1Y1CBU2 A0A1Y1CBX8 A0A1Y1CCP0 A0A1Y1CBZ5 A0A1Y1CBW7 A0A1Y1CEU1 A0A1Y1CBW1 A0A1Y1CBY5 A0A1Y1CBV9 A0A2A4J638 A0A1Y1CEV2 A0A1Y1CBT8 A0A1Y1CBV2 A0A1Y1CBV3 A0A1Y1CC06 A0A1Y1CBU6 A0A1Y1CCH5 A0A1Y1CBW2 A0A1Y1CC11 A0A1Y1CBY4 A0A1Y1CBW0 A0A1Y1CBT6 A0A336LYA0 A0A1Y1CBV7 A0A1S4GPQ0 Q7Q6Y7 A0A1Y1CEW2 A0A336M5E5 A0A1S4GPR9 A0A1S4GQD2 A0A1S4GQH6 A7UTS4 A0A336LTH9 A0A1W4VAJ8 A0A2A4J4R8 A0A0R1DZ29 A0A0P8XUJ1 A0A0Q9WL63 A0A0J9RVX2 Q2PDY9 A0A0Q5U8V3 A0A0P8ZUZ0 A0A0J9RX89 A0A0R3P3J7 M9PCN0 A0A0J9RVV6 A0A0Q9WUB4 A0A0P8YHI4 A0A0Q5UH41 A0A0J9RVW2 A0A1S4GPQ4 Q5D6W1 A0A0R1DYK3 A0A1I8NFQ3 A0A1I8NFR6 A0A1I8NFP9 A0A1I8NFS0 A0A1Y1N3S0 A0A1W4WUS7 X2JCG3 A0A1W4VNA9 A0A0R1DYI1 M9PFD4 A0A0R1DYH9 A0A0J9RW86 Q2PDZ0 A0A0J9RX61 A0A023F372 Q0GQQ8 A0A0N8P0Y2 A0A0Q9WKP9 A0A0Q5UJG1 M9NFY2 A0A0J9RX66 A0A0R1E3B3 A0A0Q5U541 A0A0J9RW88 A0A0R3P7H5 M9PFD7 A0A0Q9WKQ1 X2JGQ0 A0A0Q5UJE3 A0A0P9AKL0 A0A0J9RW83 A0A0P9AKK1 A0A0Q5U5A6 A0A0J9RVV2 M9NEB0 A0A0J9RVU6 M9PFR7
PDB
4TNW
E-value=2.79495e-39,
Score=408
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.964209
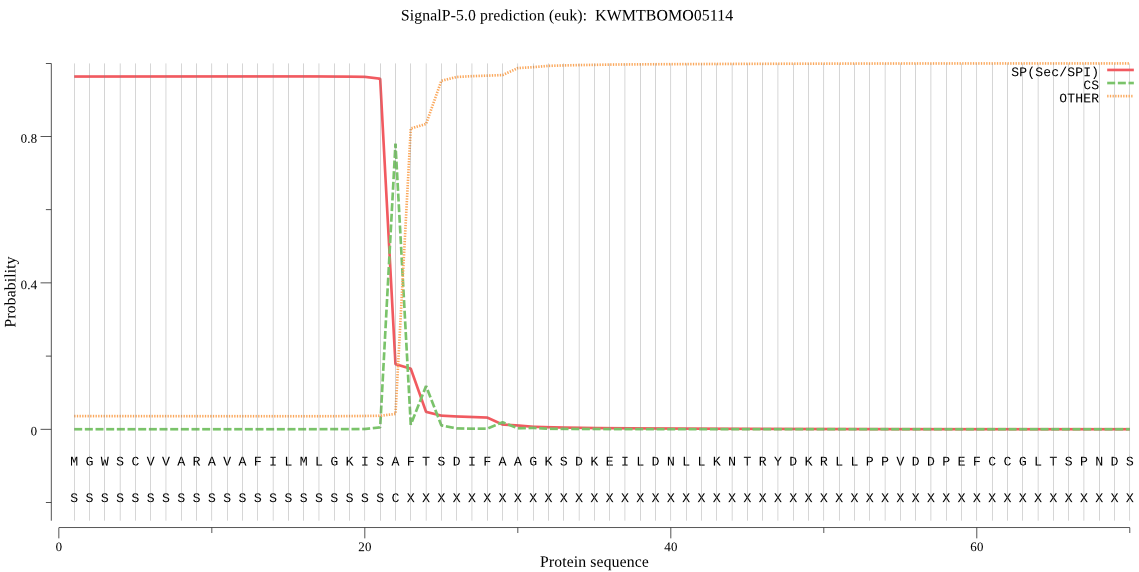
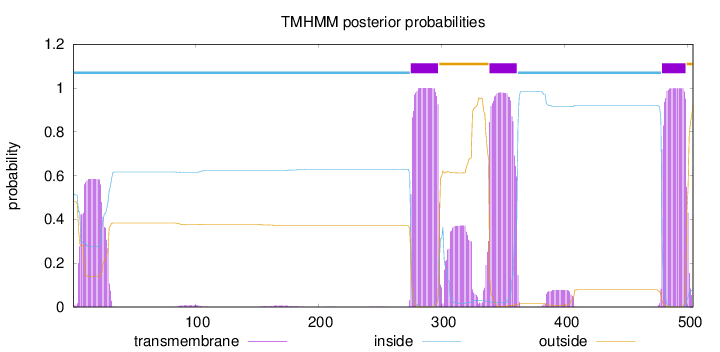
Length:
504
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
87.5421400000001
Exp number, first 60 AAs:
12.29756
Total prob of N-in:
0.51900
POSSIBLE N-term signal
sequence
inside
1 - 274
TMhelix
275 - 297
outside
298 - 338
TMhelix
339 - 361
inside
362 - 478
TMhelix
479 - 498
outside
499 - 504
Population Genetic Test Statistics
Pi
212.300855
Theta
185.117791
Tajima's D
0.245866
CLR
0.372528
CSRT
0.442777861106945
Interpretation
Uncertain
