Pre Gene Modal
BGIBMGA012505
Annotation
Thioredoxin_domain-containing_protein_[Papilio_xuthus]
Full name
MICOS complex subunit MIC60
Alternative Name
Mitofilin
Location in the cell
Cytoplasmic Reliability : 1.157 PlasmaMembrane Reliability : 1.673
Sequence
CDS
ATGAATTTTGTAATACCGTTTGTATCCCTTTTACTTTGGTGTAATAATGTAAGATCTCAAAGTTTGCTCAATGTAAGTGATGATGAACTTTTGGAACTTATAAGAGAAAAAGAAAAGCTTATTGTCTTATTCACTAAGCAAAATTGTGAGACATGTAAGAAATTGGAACAGCATGTAGAAAGTTTACAAGAAGACTTTAAGAAACACTTAAATGCTATGAGCGTTAAGACTGTAAATAGTCATTTAGCTAGACTTTACAATCCTTCAAAAGAACCAGCACTTATATTTTATCGACATGGTGTTGCACTTTTATACAGTGGTGAGGCTGATGAAAATGAAATCTATGGCTTTTTTGAGAAGAATCAAACACCAGCTGTAAAAGAGCTAACAGATAAAATATTTGAGCATTTGACACAAGCAGCTACTGGTGCAACAACAGGAGATTGGTTTGTCATGTTTTATGGGGCAGCTTGTGTTGAATGCCAAAGGCTACATGCTGTTTGGGAAAGTGTTGGAGCAACATTAAAGAGCAGAATAAATGTTGCTCGCATTGATGCAAGTTTAGCTGGTGTCAACACTGCTAAAAGATTTCATGTTGGAAAATTGCCAGCATTCCTTCTGTTTCGCCTTGGCAAAGTGTACAGATATGACTTGCCAAAAAATGATGTGAAATCTTTTGTTGCATTTGCTCAAGATTGGTATAAAAACTCTAAAGGAGAACCTGTTGCAGTTCTTGCCTCACCATTTGATGATGTGGTGAATTGGTGTGTAGAAATGCTGAAATGGGGAATAGCATTCGGTCTCAATATACTCACAGAACACCCATGGATATGGCAAATAGGAATAGCTGGTTTTGGATTGGTAGCTTTGACAGCTTTAATTGCAATTATTAAAGCAGGAAGATCAAAGCCACTAAAGGAAACAAAAAAAGATAAGAAACGTAAATAA
Protein
MNFVIPFVSLLLWCNNVRSQSLLNVSDDELLELIREKEKLIVLFTKQNCETCKKLEQHVESLQEDFKKHLNAMSVKTVNSHLARLYNPSKEPALIFYRHGVALLYSGEADENEIYGFFEKNQTPAVKELTDKIFEHLTQAATGATTGDWFVMFYGAACVECQRLHAVWESVGATLKSRINVARIDASLAGVNTAKRFHVGKLPAFLLFRLGKVYRYDLPKNDVKSFVAFAQDWYKNSKGEPVAVLASPFDDVVNWCVEMLKWGIAFGLNILTEHPWIWQIGIAGFGLVALTALIAIIKAGRSKPLKETKKDKKRK
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic60 family.
Uniprot
H9JSJ4
A0A1E1WLZ4
S4NU44
A0A194PDE6
A0A194RN86
A0A212ESH4
+ More
A0A2H1VG39 A0A2H1VHS5 A0A067RHR2 U5ES44 A0A1B0D1V2 A0A2J7QNU4 K7J4V9 A0A1L8DTZ9 A0A1L8DTY5 A0A182Y790 Q16U72 A0A182PA35 V9II76 A0A182VLR5 A0A088AS52 Q7QBY5 A0A224XNV2 A0A182MKP4 A0A2C9GR61 A0A232EP61 A0A154P768 A0A182JYI8 A0A1W7R7Z4 A0A182ISY0 R4ULE3 A0A084VJQ2 A0A0L7RHG7 A0A1Y1MJM3 A0A023EQR3 A0A1B6FW40 A0A182TD19 A0A2M3Z8F5 A0A2M3Z8C8 A0A182Q7Z4 A0A2M4ANY2 A0A2M4AP03 A0A1B6CJN6 A0A2R7WCI0 A0A2M4BUG4 A0A0P4VK18 R4G8E4 A0A1W4WE73 R4WCS0 W5JRR4 J3JV32 N6TX51 A0A1B6KMU8 A0A1Q3F8X1 A0A2M4AQY0 A0A0T6BF18 A0A2A3EU74 A0A146KPQ4 E2BMX2 E2APT1 A0A182XIC1 D6W9A7 A0A2M4ASF0 A0A026WI64 A0A182F6G5 A0A182VQA2 A0A182RCX1 E9IHM1 A0A0M9A2W4 A0A336MJM0 A0A226CXT6 A0A151WES8 E0VB17 A0A151IZE4 A0A1D2N4R9 A0A195FG00 A0A195BEM8 A0A158NF59 A0A1L8EDB3 A0A1L8ED50 B4NJV7 F4X3A6 B3P6T6 B4IJE2 A0A1W4V881 A0A0B4KHU4 B4PUZ1 B4LUV0 A0A1W4VKD2 A0A1A9VM11 A0A0P8XY41 B3M1X8 Q8SWX6 B4JQQ5 A0A1B0C4E7 A0A2S2QL25
A0A2H1VG39 A0A2H1VHS5 A0A067RHR2 U5ES44 A0A1B0D1V2 A0A2J7QNU4 K7J4V9 A0A1L8DTZ9 A0A1L8DTY5 A0A182Y790 Q16U72 A0A182PA35 V9II76 A0A182VLR5 A0A088AS52 Q7QBY5 A0A224XNV2 A0A182MKP4 A0A2C9GR61 A0A232EP61 A0A154P768 A0A182JYI8 A0A1W7R7Z4 A0A182ISY0 R4ULE3 A0A084VJQ2 A0A0L7RHG7 A0A1Y1MJM3 A0A023EQR3 A0A1B6FW40 A0A182TD19 A0A2M3Z8F5 A0A2M3Z8C8 A0A182Q7Z4 A0A2M4ANY2 A0A2M4AP03 A0A1B6CJN6 A0A2R7WCI0 A0A2M4BUG4 A0A0P4VK18 R4G8E4 A0A1W4WE73 R4WCS0 W5JRR4 J3JV32 N6TX51 A0A1B6KMU8 A0A1Q3F8X1 A0A2M4AQY0 A0A0T6BF18 A0A2A3EU74 A0A146KPQ4 E2BMX2 E2APT1 A0A182XIC1 D6W9A7 A0A2M4ASF0 A0A026WI64 A0A182F6G5 A0A182VQA2 A0A182RCX1 E9IHM1 A0A0M9A2W4 A0A336MJM0 A0A226CXT6 A0A151WES8 E0VB17 A0A151IZE4 A0A1D2N4R9 A0A195FG00 A0A195BEM8 A0A158NF59 A0A1L8EDB3 A0A1L8ED50 B4NJV7 F4X3A6 B3P6T6 B4IJE2 A0A1W4V881 A0A0B4KHU4 B4PUZ1 B4LUV0 A0A1W4VKD2 A0A1A9VM11 A0A0P8XY41 B3M1X8 Q8SWX6 B4JQQ5 A0A1B0C4E7 A0A2S2QL25
Pubmed
19121390
23622113
26354079
22118469
24845553
20075255
+ More
25244985 17510324 12364791 14747013 17210077 28648823 24438588 28004739 24945155 27129103 23691247 20920257 23761445 22516182 23537049 26823975 20798317 18362917 19820115 24508170 30249741 21282665 20566863 27289101 21347285 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356
25244985 17510324 12364791 14747013 17210077 28648823 24438588 28004739 24945155 27129103 23691247 20920257 23761445 22516182 23537049 26823975 20798317 18362917 19820115 24508170 30249741 21282665 20566863 27289101 21347285 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356
EMBL
BABH01027982
GDQN01003015
JAT88039.1
GAIX01011956
JAA80604.1
KQ459606
+ More
KPI91361.1 KQ459984 KPJ18879.1 AGBW02012813 OWR44404.1 ODYU01002391 SOQ39823.1 ODYU01002392 SOQ39824.1 KK852630 KDR19858.1 GANO01003421 JAB56450.1 AJVK01022393 NEVH01012562 PNF30252.1 AAZX01007340 GFDF01004153 JAV09931.1 GFDF01004168 JAV09916.1 CH477631 EAT38061.1 JR047201 AEY60352.1 AAAB01008859 EAA07456.4 GFTR01004928 JAW11498.1 AXCM01005074 APCN01000432 NNAY01003008 OXU20131.1 KQ434829 KZC07712.1 GEHC01000374 JAV47271.1 KC741175 AGM32999.1 ATLV01013846 KE524905 KFB38196.1 KQ414590 KOC70315.1 GEZM01031774 JAV84840.1 GAPW01002649 JAC10949.1 GECZ01015410 JAS54359.1 GGFM01004030 MBW24781.1 GGFM01004032 MBW24783.1 AXCN02000055 GGFK01009175 MBW42496.1 GGFK01009195 MBW42516.1 GEDC01023604 JAS13694.1 KK854624 PTY17394.1 GGFJ01007502 MBW56643.1 GDKW01001224 JAI55371.1 ACPB03015130 GAHY01001117 JAA76393.1 AK417113 BAN20328.1 ADMH02000291 ETN67102.1 BT127098 AEE62060.1 APGK01051641 KB741198 KB632313 ENN72971.1 ERL92167.1 GEBQ01027192 JAT12785.1 GFDL01011082 JAV23963.1 GGFK01009875 MBW43196.1 LJIG01001010 KRT85930.1 KZ288186 PBC34822.1 GDHC01020135 JAP98493.1 GL449377 EFN82954.1 GL441581 EFN64557.1 KQ971312 EEZ98470.1 GGFK01010383 MBW43704.1 KK107193 QOIP01000002 EZA55700.1 RLU26069.1 GL763289 EFZ19921.1 KQ435753 KOX76117.1 UFQT01001337 SSX30095.1 LNIX01000060 OXA37297.1 KQ983238 KYQ46291.1 DS235021 EEB10573.1 KQ980731 KYN13974.1 LJIJ01000236 ODN00055.1 KQ981606 KYN39620.1 KQ976509 KYM82642.1 ADTU01000254 GFDG01002236 JAV16563.1 GFDG01002225 JAV16574.1 CH964272 EDW83959.1 GL888609 EGI59087.1 CH954182 EDV53756.1 CH480848 EDW51133.1 AE014297 AGB96324.1 CM000160 EDW98840.1 CH940649 EDW64277.1 CH902617 KPU79621.1 EDV42238.1 AY095006 AAF56385.2 AAM11334.1 CH916372 EDV99235.1 JXJN01025433 GGMS01009266 MBY78469.1
KPI91361.1 KQ459984 KPJ18879.1 AGBW02012813 OWR44404.1 ODYU01002391 SOQ39823.1 ODYU01002392 SOQ39824.1 KK852630 KDR19858.1 GANO01003421 JAB56450.1 AJVK01022393 NEVH01012562 PNF30252.1 AAZX01007340 GFDF01004153 JAV09931.1 GFDF01004168 JAV09916.1 CH477631 EAT38061.1 JR047201 AEY60352.1 AAAB01008859 EAA07456.4 GFTR01004928 JAW11498.1 AXCM01005074 APCN01000432 NNAY01003008 OXU20131.1 KQ434829 KZC07712.1 GEHC01000374 JAV47271.1 KC741175 AGM32999.1 ATLV01013846 KE524905 KFB38196.1 KQ414590 KOC70315.1 GEZM01031774 JAV84840.1 GAPW01002649 JAC10949.1 GECZ01015410 JAS54359.1 GGFM01004030 MBW24781.1 GGFM01004032 MBW24783.1 AXCN02000055 GGFK01009175 MBW42496.1 GGFK01009195 MBW42516.1 GEDC01023604 JAS13694.1 KK854624 PTY17394.1 GGFJ01007502 MBW56643.1 GDKW01001224 JAI55371.1 ACPB03015130 GAHY01001117 JAA76393.1 AK417113 BAN20328.1 ADMH02000291 ETN67102.1 BT127098 AEE62060.1 APGK01051641 KB741198 KB632313 ENN72971.1 ERL92167.1 GEBQ01027192 JAT12785.1 GFDL01011082 JAV23963.1 GGFK01009875 MBW43196.1 LJIG01001010 KRT85930.1 KZ288186 PBC34822.1 GDHC01020135 JAP98493.1 GL449377 EFN82954.1 GL441581 EFN64557.1 KQ971312 EEZ98470.1 GGFK01010383 MBW43704.1 KK107193 QOIP01000002 EZA55700.1 RLU26069.1 GL763289 EFZ19921.1 KQ435753 KOX76117.1 UFQT01001337 SSX30095.1 LNIX01000060 OXA37297.1 KQ983238 KYQ46291.1 DS235021 EEB10573.1 KQ980731 KYN13974.1 LJIJ01000236 ODN00055.1 KQ981606 KYN39620.1 KQ976509 KYM82642.1 ADTU01000254 GFDG01002236 JAV16563.1 GFDG01002225 JAV16574.1 CH964272 EDW83959.1 GL888609 EGI59087.1 CH954182 EDV53756.1 CH480848 EDW51133.1 AE014297 AGB96324.1 CM000160 EDW98840.1 CH940649 EDW64277.1 CH902617 KPU79621.1 EDV42238.1 AY095006 AAF56385.2 AAM11334.1 CH916372 EDV99235.1 JXJN01025433 GGMS01009266 MBY78469.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000027135
UP000092462
+ More
UP000235965 UP000002358 UP000076408 UP000008820 UP000075885 UP000075903 UP000005203 UP000007062 UP000075883 UP000075840 UP000215335 UP000076502 UP000075881 UP000075880 UP000030765 UP000053825 UP000075902 UP000075886 UP000015103 UP000192223 UP000000673 UP000019118 UP000030742 UP000242457 UP000008237 UP000000311 UP000076407 UP000007266 UP000053097 UP000279307 UP000069272 UP000075920 UP000075900 UP000053105 UP000198287 UP000075809 UP000009046 UP000078492 UP000094527 UP000078541 UP000078540 UP000005205 UP000007798 UP000007755 UP000008711 UP000001292 UP000192221 UP000000803 UP000002282 UP000008792 UP000078200 UP000007801 UP000001070 UP000092460
UP000235965 UP000002358 UP000076408 UP000008820 UP000075885 UP000075903 UP000005203 UP000007062 UP000075883 UP000075840 UP000215335 UP000076502 UP000075881 UP000075880 UP000030765 UP000053825 UP000075902 UP000075886 UP000015103 UP000192223 UP000000673 UP000019118 UP000030742 UP000242457 UP000008237 UP000000311 UP000076407 UP000007266 UP000053097 UP000279307 UP000069272 UP000075920 UP000075900 UP000053105 UP000198287 UP000075809 UP000009046 UP000078492 UP000094527 UP000078541 UP000078540 UP000005205 UP000007798 UP000007755 UP000008711 UP000001292 UP000192221 UP000000803 UP000002282 UP000008792 UP000078200 UP000007801 UP000001070 UP000092460
Interpro
IPR036249
Thioredoxin-like_sf
+ More
IPR013766 Thioredoxin_domain
IPR017937 Thioredoxin_CS
IPR008554 Glutaredoxin-like
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR010893 NiFe-hyd_mat_HyaE
IPR002775 DNA/RNA-bd_Alba-like
IPR036882 Alba-like_dom_sf
IPR019133 Mt-IM_prot_Mitofilin
IPR011992 EF-hand-dom_pair
IPR013766 Thioredoxin_domain
IPR017937 Thioredoxin_CS
IPR008554 Glutaredoxin-like
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR010893 NiFe-hyd_mat_HyaE
IPR002775 DNA/RNA-bd_Alba-like
IPR036882 Alba-like_dom_sf
IPR019133 Mt-IM_prot_Mitofilin
IPR011992 EF-hand-dom_pair
Gene 3D
ProteinModelPortal
H9JSJ4
A0A1E1WLZ4
S4NU44
A0A194PDE6
A0A194RN86
A0A212ESH4
+ More
A0A2H1VG39 A0A2H1VHS5 A0A067RHR2 U5ES44 A0A1B0D1V2 A0A2J7QNU4 K7J4V9 A0A1L8DTZ9 A0A1L8DTY5 A0A182Y790 Q16U72 A0A182PA35 V9II76 A0A182VLR5 A0A088AS52 Q7QBY5 A0A224XNV2 A0A182MKP4 A0A2C9GR61 A0A232EP61 A0A154P768 A0A182JYI8 A0A1W7R7Z4 A0A182ISY0 R4ULE3 A0A084VJQ2 A0A0L7RHG7 A0A1Y1MJM3 A0A023EQR3 A0A1B6FW40 A0A182TD19 A0A2M3Z8F5 A0A2M3Z8C8 A0A182Q7Z4 A0A2M4ANY2 A0A2M4AP03 A0A1B6CJN6 A0A2R7WCI0 A0A2M4BUG4 A0A0P4VK18 R4G8E4 A0A1W4WE73 R4WCS0 W5JRR4 J3JV32 N6TX51 A0A1B6KMU8 A0A1Q3F8X1 A0A2M4AQY0 A0A0T6BF18 A0A2A3EU74 A0A146KPQ4 E2BMX2 E2APT1 A0A182XIC1 D6W9A7 A0A2M4ASF0 A0A026WI64 A0A182F6G5 A0A182VQA2 A0A182RCX1 E9IHM1 A0A0M9A2W4 A0A336MJM0 A0A226CXT6 A0A151WES8 E0VB17 A0A151IZE4 A0A1D2N4R9 A0A195FG00 A0A195BEM8 A0A158NF59 A0A1L8EDB3 A0A1L8ED50 B4NJV7 F4X3A6 B3P6T6 B4IJE2 A0A1W4V881 A0A0B4KHU4 B4PUZ1 B4LUV0 A0A1W4VKD2 A0A1A9VM11 A0A0P8XY41 B3M1X8 Q8SWX6 B4JQQ5 A0A1B0C4E7 A0A2S2QL25
A0A2H1VG39 A0A2H1VHS5 A0A067RHR2 U5ES44 A0A1B0D1V2 A0A2J7QNU4 K7J4V9 A0A1L8DTZ9 A0A1L8DTY5 A0A182Y790 Q16U72 A0A182PA35 V9II76 A0A182VLR5 A0A088AS52 Q7QBY5 A0A224XNV2 A0A182MKP4 A0A2C9GR61 A0A232EP61 A0A154P768 A0A182JYI8 A0A1W7R7Z4 A0A182ISY0 R4ULE3 A0A084VJQ2 A0A0L7RHG7 A0A1Y1MJM3 A0A023EQR3 A0A1B6FW40 A0A182TD19 A0A2M3Z8F5 A0A2M3Z8C8 A0A182Q7Z4 A0A2M4ANY2 A0A2M4AP03 A0A1B6CJN6 A0A2R7WCI0 A0A2M4BUG4 A0A0P4VK18 R4G8E4 A0A1W4WE73 R4WCS0 W5JRR4 J3JV32 N6TX51 A0A1B6KMU8 A0A1Q3F8X1 A0A2M4AQY0 A0A0T6BF18 A0A2A3EU74 A0A146KPQ4 E2BMX2 E2APT1 A0A182XIC1 D6W9A7 A0A2M4ASF0 A0A026WI64 A0A182F6G5 A0A182VQA2 A0A182RCX1 E9IHM1 A0A0M9A2W4 A0A336MJM0 A0A226CXT6 A0A151WES8 E0VB17 A0A151IZE4 A0A1D2N4R9 A0A195FG00 A0A195BEM8 A0A158NF59 A0A1L8EDB3 A0A1L8ED50 B4NJV7 F4X3A6 B3P6T6 B4IJE2 A0A1W4V881 A0A0B4KHU4 B4PUZ1 B4LUV0 A0A1W4VKD2 A0A1A9VM11 A0A0P8XY41 B3M1X8 Q8SWX6 B4JQQ5 A0A1B0C4E7 A0A2S2QL25
PDB
3IDV
E-value=2.30975e-07,
Score=130
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
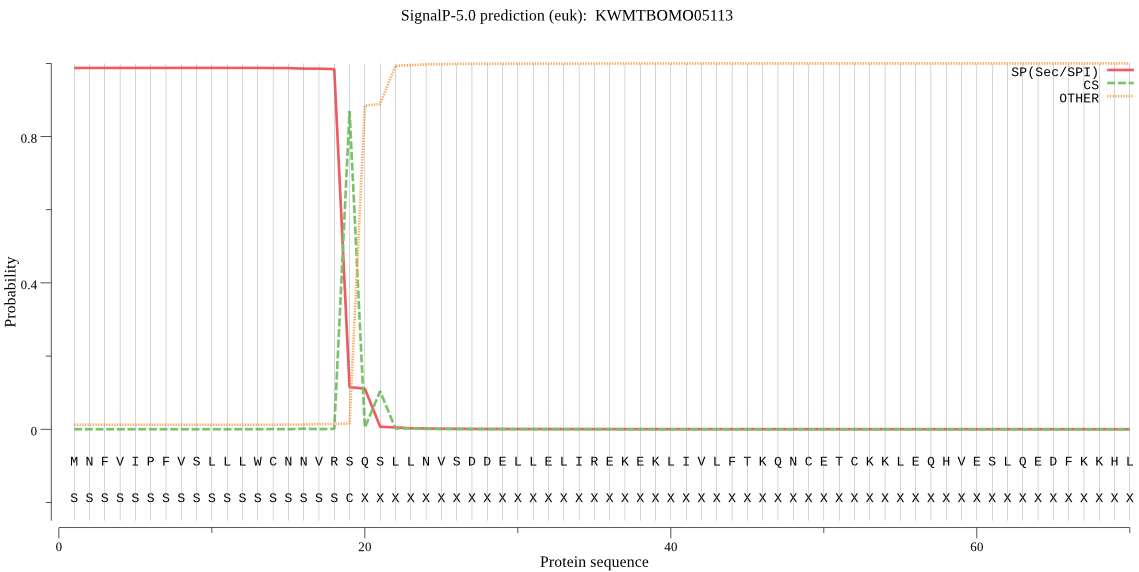
SignalP
Position: 1 - 19,
Likelihood: 0.987028
Length:
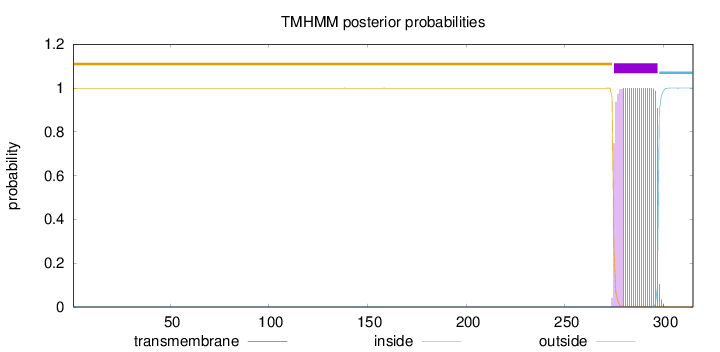
315
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.74913
Exp number, first 60 AAs:
0.001
Total prob of N-in:
0.00059
outside
1 - 274
TMhelix
275 - 297
inside
298 - 315
Population Genetic Test Statistics
Pi
264.287208
Theta
200.42501
Tajima's D
1.089802
CLR
0.007623
CSRT
0.689115544222789
Interpretation
Uncertain
