Gene
KWMTBOMO05111 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012506
Annotation
PREDICTED:_uncharacterized_protein_LOC106134724_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.808
Sequence
CDS
ATGGCCGAAGTTGAAGAAAAGGTGATAATGACACCTAAATGTAAAACGGCCAATTCCACAACATTGGTAGTTGAAAGGAAAGCTGTAGAACCAGAAGCAAGTGACAAAATACATATAGCTGGTGGTGATCACACTGGTATTATTATCAACAAGCAAGAAACTTATGAAAATGGCGTTTCCGAGCCTTGTCATGCACAATTGGAATTCTATGTTTATTTGGTATCGGGAGCTACTGGAACCCACACACGTGAAGCACGTGCTCTAAGGTTCTGGTTTAAACCTCAGATGACTCCAAACGAAAGGCCTTATGAAGCCCAGGCCTTTTTCAGAGAACTTGTCAGTCCTCAAGATTTTCCTAAAGATTATGTAGGATACATAAAAAAAATAATGAAATTAATGCAACACAAATATCAACAACTGAAAATGCTTGAAGTAGAATTAAGACAAGAAGGATATGCATCACCACCACCAGCATACATCGATGACAGTATAATAAATCAAACACCACTAATATCAGAACAAAGAGTGCTGGATATGATAGAAAACGCATATCCCAACCCCTTATCTGTGGATGATTTTGTCTCTGCCGCCAAATGGTCGAAAGCAGACGTGAAGGATGCATTGGAATCTTTAGAAGAAAAAGGCCTCACTCGTGCTATGTCCGAAGGTGTTTATGTTCGTCAACATAGCGTAGATACACAGGTAGTAAAACAGATGCCAACGTTGAGTTCGTCACGTCAACCCAGTATTGCAGTCATAACAGCGTTGTATTGTGAAAAACAAGCCGTTGATGCAATGATGGACAATCAAGAAACCTACGTACGTTATACCACTGTTGGAGAATCGAATGTTTATACATTGGGAACAATCGGTGCTCATCGAGTTGTCACTACTAAACTACCTTCCGTGGGCCGTACCAGAGAAGCAATGACTGCAGCCGGCAATACCACTACAAGACTTCTTGGAATCTTTCAAAAGGTCGAATACGTTTTCCTAGTCGGCGTGGGCGGAGGAGTACCTCACTATACGGATTATTCGAAACATGTGCGTCTAGGTGACGTGGTTGTTTCCACGCCGGCGCCAGACTTGCCAGACAAATACGTATACGTTTTCTGTGAAAAAGCCGAGAAGAACGCCAAAGGGGGTTGGGATTTCGAAGTGAAGCCATACAAGCCAGTTAATTTCTTATTGCAAGATATCGCTGTGCGTCTCACCAAGTCAAAGGCCTCATGGGATTTGTACGTCAAAGAAGGACTCGATCGTCTCAGAGGAGTCCCGGGACGATGGGAGCCCCCGGAGAGCAACACCGACAGGTTGTGCGTCGAAGTCGGCAATGGAGCTTACATTGAAGTATCACATCCAGCGCCACAAGGAGATCAAACTGACACGCGGGACGGGTTGCGCGTGACGTGCGCGCCCGTGGCGGGCGGGCGCGTGGTATCGGCGGCGGGCGAGGGGCGGGCCGCCTTCGCCGCCCACACGCGCGCGCTCGCCTACGACTGCGAGCTCGACGCCGTGCTCGACAGCGTCGTCGGCAACTGCAAGGACTGCTTCATATCGCTGCGAGGTATATCGGACTATCGCGACGGCACCCGTGGGAAGGACTGGCAGCCCCACGCTGCACTATGCGCTGCTGCAGTCATGAAAGCCATAATCTGCGCAATAGACCCACCTCCTTCTGAATAA
Protein
MAEVEEKVIMTPKCKTANSTTLVVERKAVEPEASDKIHIAGGDHTGIIINKQETYENGVSEPCHAQLEFYVYLVSGATGTHTREARALRFWFKPQMTPNERPYEAQAFFRELVSPQDFPKDYVGYIKKIMKLMQHKYQQLKMLEVELRQEGYASPPPAYIDDSIINQTPLISEQRVLDMIENAYPNPLSVDDFVSAAKWSKADVKDALESLEEKGLTRAMSEGVYVRQHSVDTQVVKQMPTLSSSRQPSIAVITALYCEKQAVDAMMDNQETYVRYTTVGESNVYTLGTIGAHRVVTTKLPSVGRTREAMTAAGNTTTRLLGIFQKVEYVFLVGVGGGVPHYTDYSKHVRLGDVVVSTPAPDLPDKYVYVFCEKAEKNAKGGWDFEVKPYKPVNFLLQDIAVRLTKSKASWDLYVKEGLDRLRGVPGRWEPPESNTDRLCVEVGNGAYIEVSHPAPQGDQTDTRDGLRVTCAPVAGGRVVSAAGEGRAAFAAHTRALAYDCELDAVLDSVVGNCKDCFISLRGISDYRDGTRGKDWQPHAALCAAAVMKAIICAIDPPPSE
Summary
Uniprot
A0A194PF82
A0A212ESD5
A0A1E1WAH8
A0A1Y1LTX9
A0A1Y1LSW4
A0A1Y1LNX8
+ More
A0A1Y1LQZ3 D6W9B6 E2B7N4 A0A0C9RIQ7 A0A026VT37 A0A3L8D3A9 K7IWL1 A0A0C9PLH9 A0A2A3E9Z7 A0A0L7QZJ2 E9ILU1 E1ZZ32 A0A310SAK3 F4WAK3 A0A158N9C6 A0A0A9XR35 A0A0P4W7Y8 A0A2J7PT55 A0A067R4H4 A0A151XFZ3 A0A151J5A3 A0A1W4V307 A0A0R1ECW1 A0A0Q5TJ41 Q8MRM6 A0A0N8P0K4 A0A0Q9WLN0 A0A1L8E683 A0A0R3NZY9 A0A088A8E1 A0A195F8P1 T1P7X6 A0A3B0KTI5 A0A0P4VN46 A0A151I3W8 A0A1B6L3H2 A0A0Q9WNI8 A0A336KWX4 U4U285 A0A1W4VGK8 A0A1W4V2Y4 A0A0Q5TE54 A0A0R1EHV4 A0A0P8YEN9 A0A1I8MFP7 A0A0P9C180 A0A1J1IVL2 A0A0Q9WMX5 A0A0R3P5Z0 A0A1L8E6A7 A0A0R3P5D0 U5ECT1 A0A0P6IX67 A0A3B0K7B9 A0A0Q9WU78 A0A3B0K5K8 A0A1A9W338 A0A1L8DCN3 A0A1I8MFQ9 A0A0Q9WZB7 A0A069DV69 A0A023F0Y5 A0A023EVN6 B3NVF6 F5HJL6 A0A182XBU2 B4Q095 Q9W3D6 A0A1Q3FG91 A0A1W4V2X4 A0A1Q3FGE2 A0A1A9VBX2 B3N0F2 B4R6K1 B4M2B2 A0A336MIC7 A0A3B0KJ53 A0A2M4CRM3 A0A2M4CT26 A0A034V6F4 Q29HF4 W8AHY6 A0A224XM39 A0A1I8MFP8 A0A1J1IYM6 A0A0K8V904 B4L1V7 A0A1J1IXF0 B4GXT5 A0A0P6IWF0 A0A1L8DCP8 B4MSD2
A0A1Y1LQZ3 D6W9B6 E2B7N4 A0A0C9RIQ7 A0A026VT37 A0A3L8D3A9 K7IWL1 A0A0C9PLH9 A0A2A3E9Z7 A0A0L7QZJ2 E9ILU1 E1ZZ32 A0A310SAK3 F4WAK3 A0A158N9C6 A0A0A9XR35 A0A0P4W7Y8 A0A2J7PT55 A0A067R4H4 A0A151XFZ3 A0A151J5A3 A0A1W4V307 A0A0R1ECW1 A0A0Q5TJ41 Q8MRM6 A0A0N8P0K4 A0A0Q9WLN0 A0A1L8E683 A0A0R3NZY9 A0A088A8E1 A0A195F8P1 T1P7X6 A0A3B0KTI5 A0A0P4VN46 A0A151I3W8 A0A1B6L3H2 A0A0Q9WNI8 A0A336KWX4 U4U285 A0A1W4VGK8 A0A1W4V2Y4 A0A0Q5TE54 A0A0R1EHV4 A0A0P8YEN9 A0A1I8MFP7 A0A0P9C180 A0A1J1IVL2 A0A0Q9WMX5 A0A0R3P5Z0 A0A1L8E6A7 A0A0R3P5D0 U5ECT1 A0A0P6IX67 A0A3B0K7B9 A0A0Q9WU78 A0A3B0K5K8 A0A1A9W338 A0A1L8DCN3 A0A1I8MFQ9 A0A0Q9WZB7 A0A069DV69 A0A023F0Y5 A0A023EVN6 B3NVF6 F5HJL6 A0A182XBU2 B4Q095 Q9W3D6 A0A1Q3FG91 A0A1W4V2X4 A0A1Q3FGE2 A0A1A9VBX2 B3N0F2 B4R6K1 B4M2B2 A0A336MIC7 A0A3B0KJ53 A0A2M4CRM3 A0A2M4CT26 A0A034V6F4 Q29HF4 W8AHY6 A0A224XM39 A0A1I8MFP8 A0A1J1IYM6 A0A0K8V904 B4L1V7 A0A1J1IXF0 B4GXT5 A0A0P6IWF0 A0A1L8DCP8 B4MSD2
Pubmed
26354079
22118469
28004739
18362917
19820115
20798317
+ More
24508170 30249741 20075255 21282665 21719571 21347285 25401762 24845553 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25315136 27129103 23537049 18057021 26999592 26334808 25474469 24945155 12364791 14747013 17210077 25348373 23185243 24495485
24508170 30249741 20075255 21282665 21719571 21347285 25401762 24845553 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25315136 27129103 23537049 18057021 26999592 26334808 25474469 24945155 12364791 14747013 17210077 25348373 23185243 24495485
EMBL
KQ459606
KPI91359.1
AGBW02012813
OWR44405.1
GDQN01007052
GDQN01005046
+ More
JAT84002.1 JAT86008.1 GEZM01050777 JAV75385.1 GEZM01050782 JAV75380.1 GEZM01050778 JAV75384.1 GEZM01050772 JAV75391.1 KQ971312 EEZ98475.1 GL446193 EFN88312.1 GBYB01013127 JAG82894.1 KK108821 EZA46626.1 QOIP01000014 RLU14726.1 GBYB01001928 JAG71695.1 KZ288325 PBC28019.1 KQ414680 KOC64018.1 GL764129 EFZ18325.1 GL435281 EFN73551.1 KQ781845 OAD51894.1 GL888050 EGI68761.1 ADTU01009383 ADTU01009384 GBHO01041311 GBHO01041310 GBHO01020448 GBHO01020432 GBRD01018176 JAG02293.1 JAG02294.1 JAG23156.1 JAG23172.1 JAG47651.1 GDRN01083801 JAI61617.1 NEVH01021919 PNF19521.1 KK852901 KDR14122.1 KQ982174 KYQ59296.1 KQ980026 KYN18202.1 CM000162 KRK06654.1 CH954180 KQS29942.1 AY119523 AE014298 BT023715 AAM50177.1 AAS65291.1 AAY85115.1 CH902640 KPU77372.1 CH940651 KRF82433.1 GFDF01000019 JAV14065.1 CH379064 KRT06578.1 KQ981727 KYN36746.1 KA644731 AFP59360.1 OUUW01000015 SPP88571.1 GDKW01000560 JAI56035.1 KQ976484 KYM83685.1 GEBQ01021714 JAT18263.1 CH933810 KRF93803.1 UFQS01001256 UFQT01001256 SSX09979.1 SSX29701.1 KB631924 ERL87182.1 KQS29941.1 KRK06650.1 KPU77373.1 KPU77374.1 CVRI01000063 CRL04253.1 KRF82431.1 KRF82432.1 KRT06582.1 GFDF01000022 JAV14062.1 KRT06579.1 GANO01004988 JAB54883.1 GDUN01000487 JAN95432.1 SPP88572.1 KRF93804.1 SPP88573.1 GFDF01009861 JAV04223.1 KRF93802.1 GBGD01000936 JAC87953.1 GBBI01003562 JAC15150.1 GAPW01000476 JAC13122.1 EDV46348.1 KQS29939.1 KQS29940.1 KQS29943.1 KQS29944.1 AAAB01008846 EGK96477.1 EDX02232.2 KRK06649.1 KRK06651.1 KRK06652.1 KRK06653.1 AAF46394.1 AAN09237.1 GFDL01008475 JAV26570.1 GFDL01008434 JAV26611.1 EDV38356.1 KPU77375.1 CM000366 EDX17431.1 EDW65816.1 SSX09978.1 SSX29700.1 SPP88570.1 GGFL01003808 MBW67986.1 GGFL01003810 MBW67988.1 GAKP01020853 JAC38099.1 EAL31804.3 KRT06580.1 KRT06581.1 GAMC01018460 JAB88095.1 GFTR01007335 JAW09091.1 CRL04254.1 GDHF01016880 JAI35434.1 EDW06760.2 CRL04252.1 CH479196 EDW27562.1 GDUN01000488 JAN95431.1 GFDF01009846 JAV04238.1 CH963851 EDW75021.2
JAT84002.1 JAT86008.1 GEZM01050777 JAV75385.1 GEZM01050782 JAV75380.1 GEZM01050778 JAV75384.1 GEZM01050772 JAV75391.1 KQ971312 EEZ98475.1 GL446193 EFN88312.1 GBYB01013127 JAG82894.1 KK108821 EZA46626.1 QOIP01000014 RLU14726.1 GBYB01001928 JAG71695.1 KZ288325 PBC28019.1 KQ414680 KOC64018.1 GL764129 EFZ18325.1 GL435281 EFN73551.1 KQ781845 OAD51894.1 GL888050 EGI68761.1 ADTU01009383 ADTU01009384 GBHO01041311 GBHO01041310 GBHO01020448 GBHO01020432 GBRD01018176 JAG02293.1 JAG02294.1 JAG23156.1 JAG23172.1 JAG47651.1 GDRN01083801 JAI61617.1 NEVH01021919 PNF19521.1 KK852901 KDR14122.1 KQ982174 KYQ59296.1 KQ980026 KYN18202.1 CM000162 KRK06654.1 CH954180 KQS29942.1 AY119523 AE014298 BT023715 AAM50177.1 AAS65291.1 AAY85115.1 CH902640 KPU77372.1 CH940651 KRF82433.1 GFDF01000019 JAV14065.1 CH379064 KRT06578.1 KQ981727 KYN36746.1 KA644731 AFP59360.1 OUUW01000015 SPP88571.1 GDKW01000560 JAI56035.1 KQ976484 KYM83685.1 GEBQ01021714 JAT18263.1 CH933810 KRF93803.1 UFQS01001256 UFQT01001256 SSX09979.1 SSX29701.1 KB631924 ERL87182.1 KQS29941.1 KRK06650.1 KPU77373.1 KPU77374.1 CVRI01000063 CRL04253.1 KRF82431.1 KRF82432.1 KRT06582.1 GFDF01000022 JAV14062.1 KRT06579.1 GANO01004988 JAB54883.1 GDUN01000487 JAN95432.1 SPP88572.1 KRF93804.1 SPP88573.1 GFDF01009861 JAV04223.1 KRF93802.1 GBGD01000936 JAC87953.1 GBBI01003562 JAC15150.1 GAPW01000476 JAC13122.1 EDV46348.1 KQS29939.1 KQS29940.1 KQS29943.1 KQS29944.1 AAAB01008846 EGK96477.1 EDX02232.2 KRK06649.1 KRK06651.1 KRK06652.1 KRK06653.1 AAF46394.1 AAN09237.1 GFDL01008475 JAV26570.1 GFDL01008434 JAV26611.1 EDV38356.1 KPU77375.1 CM000366 EDX17431.1 EDW65816.1 SSX09978.1 SSX29700.1 SPP88570.1 GGFL01003808 MBW67986.1 GGFL01003810 MBW67988.1 GAKP01020853 JAC38099.1 EAL31804.3 KRT06580.1 KRT06581.1 GAMC01018460 JAB88095.1 GFTR01007335 JAW09091.1 CRL04254.1 GDHF01016880 JAI35434.1 EDW06760.2 CRL04252.1 CH479196 EDW27562.1 GDUN01000488 JAN95431.1 GFDF01009846 JAV04238.1 CH963851 EDW75021.2
Proteomes
UP000053268
UP000007151
UP000007266
UP000008237
UP000053097
UP000279307
+ More
UP000002358 UP000242457 UP000053825 UP000000311 UP000007755 UP000005205 UP000235965 UP000027135 UP000075809 UP000078492 UP000192221 UP000002282 UP000008711 UP000000803 UP000007801 UP000008792 UP000001819 UP000005203 UP000078541 UP000095301 UP000268350 UP000078540 UP000009192 UP000030742 UP000183832 UP000091820 UP000007062 UP000076407 UP000078200 UP000000304 UP000008744 UP000007798
UP000002358 UP000242457 UP000053825 UP000000311 UP000007755 UP000005205 UP000235965 UP000027135 UP000075809 UP000078492 UP000192221 UP000002282 UP000008711 UP000000803 UP000007801 UP000008792 UP000001819 UP000005203 UP000078541 UP000095301 UP000268350 UP000078540 UP000009192 UP000030742 UP000183832 UP000091820 UP000007062 UP000076407 UP000078200 UP000000304 UP000008744 UP000007798
Pfam
PF01048 PNP_UDP_1
Interpro
Gene 3D
ProteinModelPortal
A0A194PF82
A0A212ESD5
A0A1E1WAH8
A0A1Y1LTX9
A0A1Y1LSW4
A0A1Y1LNX8
+ More
A0A1Y1LQZ3 D6W9B6 E2B7N4 A0A0C9RIQ7 A0A026VT37 A0A3L8D3A9 K7IWL1 A0A0C9PLH9 A0A2A3E9Z7 A0A0L7QZJ2 E9ILU1 E1ZZ32 A0A310SAK3 F4WAK3 A0A158N9C6 A0A0A9XR35 A0A0P4W7Y8 A0A2J7PT55 A0A067R4H4 A0A151XFZ3 A0A151J5A3 A0A1W4V307 A0A0R1ECW1 A0A0Q5TJ41 Q8MRM6 A0A0N8P0K4 A0A0Q9WLN0 A0A1L8E683 A0A0R3NZY9 A0A088A8E1 A0A195F8P1 T1P7X6 A0A3B0KTI5 A0A0P4VN46 A0A151I3W8 A0A1B6L3H2 A0A0Q9WNI8 A0A336KWX4 U4U285 A0A1W4VGK8 A0A1W4V2Y4 A0A0Q5TE54 A0A0R1EHV4 A0A0P8YEN9 A0A1I8MFP7 A0A0P9C180 A0A1J1IVL2 A0A0Q9WMX5 A0A0R3P5Z0 A0A1L8E6A7 A0A0R3P5D0 U5ECT1 A0A0P6IX67 A0A3B0K7B9 A0A0Q9WU78 A0A3B0K5K8 A0A1A9W338 A0A1L8DCN3 A0A1I8MFQ9 A0A0Q9WZB7 A0A069DV69 A0A023F0Y5 A0A023EVN6 B3NVF6 F5HJL6 A0A182XBU2 B4Q095 Q9W3D6 A0A1Q3FG91 A0A1W4V2X4 A0A1Q3FGE2 A0A1A9VBX2 B3N0F2 B4R6K1 B4M2B2 A0A336MIC7 A0A3B0KJ53 A0A2M4CRM3 A0A2M4CT26 A0A034V6F4 Q29HF4 W8AHY6 A0A224XM39 A0A1I8MFP8 A0A1J1IYM6 A0A0K8V904 B4L1V7 A0A1J1IXF0 B4GXT5 A0A0P6IWF0 A0A1L8DCP8 B4MSD2
A0A1Y1LQZ3 D6W9B6 E2B7N4 A0A0C9RIQ7 A0A026VT37 A0A3L8D3A9 K7IWL1 A0A0C9PLH9 A0A2A3E9Z7 A0A0L7QZJ2 E9ILU1 E1ZZ32 A0A310SAK3 F4WAK3 A0A158N9C6 A0A0A9XR35 A0A0P4W7Y8 A0A2J7PT55 A0A067R4H4 A0A151XFZ3 A0A151J5A3 A0A1W4V307 A0A0R1ECW1 A0A0Q5TJ41 Q8MRM6 A0A0N8P0K4 A0A0Q9WLN0 A0A1L8E683 A0A0R3NZY9 A0A088A8E1 A0A195F8P1 T1P7X6 A0A3B0KTI5 A0A0P4VN46 A0A151I3W8 A0A1B6L3H2 A0A0Q9WNI8 A0A336KWX4 U4U285 A0A1W4VGK8 A0A1W4V2Y4 A0A0Q5TE54 A0A0R1EHV4 A0A0P8YEN9 A0A1I8MFP7 A0A0P9C180 A0A1J1IVL2 A0A0Q9WMX5 A0A0R3P5Z0 A0A1L8E6A7 A0A0R3P5D0 U5ECT1 A0A0P6IX67 A0A3B0K7B9 A0A0Q9WU78 A0A3B0K5K8 A0A1A9W338 A0A1L8DCN3 A0A1I8MFQ9 A0A0Q9WZB7 A0A069DV69 A0A023F0Y5 A0A023EVN6 B3NVF6 F5HJL6 A0A182XBU2 B4Q095 Q9W3D6 A0A1Q3FG91 A0A1W4V2X4 A0A1Q3FGE2 A0A1A9VBX2 B3N0F2 B4R6K1 B4M2B2 A0A336MIC7 A0A3B0KJ53 A0A2M4CRM3 A0A2M4CT26 A0A034V6F4 Q29HF4 W8AHY6 A0A224XM39 A0A1I8MFP8 A0A1J1IYM6 A0A0K8V904 B4L1V7 A0A1J1IXF0 B4GXT5 A0A0P6IWF0 A0A1L8DCP8 B4MSD2
Ontologies
GO
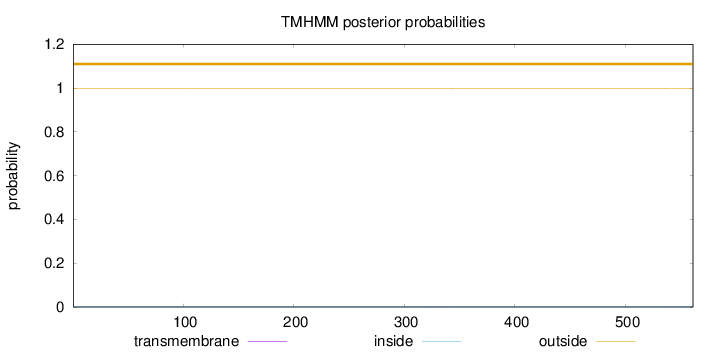
Topology
Length:
561
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03184
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00191
outside
1 - 561
Population Genetic Test Statistics
Pi
227.41917
Theta
177.86772
Tajima's D
0.807493
CLR
0.383851
CSRT
0.603369831508425
Interpretation
Uncertain
