Pre Gene Modal
BGIBMGA012459
Annotation
TBP-associated_factor_13_isoform_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.631
Sequence
CDS
ATGGCCACTCCCACTCCTGATTCGCAAGATAACTTTGACCAGTTTGATGAAGAAGAAGAGCAACAGTTAGGTGGAACTACATCTGGTCGTAAAAGACTTTTTAGTAAGGAATTGAGATGTATGATGTATGGATTTGGTGATGATCAAAACCCGTATACTGAAAGTGTCGATTTTTTAGAAGACTTAGTTATTGAATTTATAACAGAAACAACACATAGGGCAATGGAAGTTGGTAGAACAGGAAGAGTCCAAGTAGAAGACATAATTTTCTTAGTTCGTAAAGATGCACGAAAGTATGCACGTGTTAAAGACCTTCTTACAATGAATGAGGAACTTAAGAAAGCTAGAAAAGCTTTTGATGAAGTAAAATATGTTGAGGACCAACAGTAA
Protein
MATPTPDSQDNFDQFDEEEEQQLGGTTSGRKRLFSKELRCMMYGFGDDQNPYTESVDFLEDLVIEFITETTHRAMEVGRTGRVQVEDIIFLVRKDARKYARVKDLLTMNEELKKARKAFDEVKYVEDQQ
Summary
Uniprot
Q1HQ71
H9JSE8
Q1HQ72
S4NLJ7
A0A2A4IVC1
A0A2A4ITT2
+ More
A0A1E1W7Y8 A0A3S2NBV3 A0A2H1VUF6 A0A1E1WLG1 A0A2A4IUL8 A0A0L7KQ66 I4DS46 A0A0K8TPR7 A0A194PDM5 A0A194RPY7 A0A212ESD1 A0A067QVU2 A0A232F8B8 K7IR23 R4V1D4 A0A2P8XB90 A0A0C9QW43 B0W7M4 A0A1B0C904 A0A1B0AH71 A0A0C9RI76 U5ENY6 Q17JG3 A0A023EEG2 A0A0K8UXD3 A0A182RC81 A0A182Y020 A0A1B0DM14 A0A1B0G2F6 A0A1A9UD09 A0A182MBD1 A0A182STT5 A0A151WPH5 A0A034UZ26 T1E8S3 A0A2M4AX09 A0A2M4C319 F4X177 A0A158NFQ2 A0A1B6DKU2 A0A084VND8 W8BZX5 A0A195BDM0 A0A151JY37 A0A195CW55 E2C4H7 A0A1L8DHA1 A0A0P4XBQ4 A0A1L8EDN2 A0A2M4C3R6 A0A1L8DH42 A0A1I8M6K8 A0A1I8QB52 A0A0P5TCC7 A0A0L7RFX2 A0A1L8EBM8 A0A0J7L4M6 E9GLB8 A0A1W4VXW9 E2AU31 W5JK90 A0A2A3E4N3 A0A087ZX98 A0A1Y1N4V2 E0W312 A0A182TSS8 Q7Q3U1 A0A1A9WHG0 A0A0L0C4W3 A0A164XUE7 Q29KL1 B4GT64 A0A1A9YRT3 A0A182GSK9 A0A0P4X1R9 A0A3B0JV47 B4JQ95 A0A3L8E3T7 B4KEV9 B3MKW0 A0A0P5WUM4 A0A1B0AKN3 A0A1B0C698 A0A026WDJ0 A0A0M4EDP2 B4NS08 Q9VIP1 B3NL01 B4PBI0 B4M9M3 B4I636 B4N7S0 A0A0T6BHD0 A0A087UXG5
A0A1E1W7Y8 A0A3S2NBV3 A0A2H1VUF6 A0A1E1WLG1 A0A2A4IUL8 A0A0L7KQ66 I4DS46 A0A0K8TPR7 A0A194PDM5 A0A194RPY7 A0A212ESD1 A0A067QVU2 A0A232F8B8 K7IR23 R4V1D4 A0A2P8XB90 A0A0C9QW43 B0W7M4 A0A1B0C904 A0A1B0AH71 A0A0C9RI76 U5ENY6 Q17JG3 A0A023EEG2 A0A0K8UXD3 A0A182RC81 A0A182Y020 A0A1B0DM14 A0A1B0G2F6 A0A1A9UD09 A0A182MBD1 A0A182STT5 A0A151WPH5 A0A034UZ26 T1E8S3 A0A2M4AX09 A0A2M4C319 F4X177 A0A158NFQ2 A0A1B6DKU2 A0A084VND8 W8BZX5 A0A195BDM0 A0A151JY37 A0A195CW55 E2C4H7 A0A1L8DHA1 A0A0P4XBQ4 A0A1L8EDN2 A0A2M4C3R6 A0A1L8DH42 A0A1I8M6K8 A0A1I8QB52 A0A0P5TCC7 A0A0L7RFX2 A0A1L8EBM8 A0A0J7L4M6 E9GLB8 A0A1W4VXW9 E2AU31 W5JK90 A0A2A3E4N3 A0A087ZX98 A0A1Y1N4V2 E0W312 A0A182TSS8 Q7Q3U1 A0A1A9WHG0 A0A0L0C4W3 A0A164XUE7 Q29KL1 B4GT64 A0A1A9YRT3 A0A182GSK9 A0A0P4X1R9 A0A3B0JV47 B4JQ95 A0A3L8E3T7 B4KEV9 B3MKW0 A0A0P5WUM4 A0A1B0AKN3 A0A1B0C698 A0A026WDJ0 A0A0M4EDP2 B4NS08 Q9VIP1 B3NL01 B4PBI0 B4M9M3 B4I636 B4N7S0 A0A0T6BHD0 A0A087UXG5
Pubmed
19121390
23622113
26227816
22651552
26369729
26354079
+ More
22118469 24845553 28648823 20075255 29403074 17510324 24945155 25244985 25348373 21719571 21347285 24438588 24495485 20798317 25315136 21292972 20920257 23761445 28004739 20566863 12364791 14747013 17210077 26108605 15632085 17994087 26483478 30249741 24508170 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
22118469 24845553 28648823 20075255 29403074 17510324 24945155 25244985 25348373 21719571 21347285 24438588 24495485 20798317 25315136 21292972 20920257 23761445 28004739 20566863 12364791 14747013 17210077 26108605 15632085 17994087 26483478 30249741 24508170 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
DQ443181
ABF51270.1
BABH01027984
DQ443180
ABF51269.1
GAIX01014666
+ More
JAA77894.1 NWSH01007107 PCG63090.1 PCG63089.1 GDQN01008728 GDQN01007990 JAT82326.1 JAT83064.1 RSAL01000257 RVE43411.1 ODYU01004478 SOQ44386.1 GDQN01003239 JAT87815.1 PCG63088.1 JTDY01007558 KOB65114.1 AK405418 BAM20736.1 GDAI01001713 JAI15890.1 KQ459606 KPI91357.1 KQ459896 KPJ19469.1 AGBW02012813 OWR44406.1 KK852898 KDR14167.1 NNAY01000669 OXU27084.1 KC571943 AGM32442.1 PYGN01004758 PYGN01000725 PSN29271.1 PSN41633.1 GBYB01007959 JAG77726.1 DS231854 EDS38059.1 AJWK01001675 GBYB01007960 JAG77727.1 GANO01000393 JAB59478.1 CH477232 EAT46877.1 GAPW01005841 GAPW01005840 JAC07757.1 GDHF01021284 JAI31030.1 AJVK01006816 CCAG010014651 AXCM01001558 KQ982893 KYQ49595.1 GAKP01023419 JAC35539.1 GAMD01002508 JAA99082.1 GGFK01011989 MBW45310.1 GGFJ01010553 MBW59694.1 GL888528 EGI59749.1 ADTU01014508 GEDC01011023 JAS26275.1 ATLV01014737 KE524984 KFB39482.1 GAMC01007656 GAMC01007654 JAB98901.1 KQ976514 KYM82280.1 KQ981494 KYN40933.1 KQ977279 KYN04389.1 GL452471 EFN77161.1 GFDF01008319 JAV05765.1 GDIP01246519 JAI76882.1 GFDG01001969 JAV16830.1 GGFJ01010829 MBW59970.1 GFDF01008318 JAV05766.1 GDIP01129461 JAL74253.1 KQ414605 KOC69636.1 GFDG01002774 JAV16025.1 LBMM01000784 KMQ97508.1 GL732551 EFX79482.1 GL442767 EFN63074.1 ADMH02000877 ETN64792.1 KZ288376 PBC26665.1 GEZM01015027 JAV91920.1 DS235881 EEB20018.1 AAAB01008964 EAA12355.3 JRES01000914 KNC27281.1 LRGB01000944 KZS14579.1 CH379061 EAL33163.1 CH479189 EDW25573.1 JXUM01017001 KQ560472 KXJ82233.1 GDIP01248200 JAI75201.1 OUUW01000010 SPP85977.1 CH916372 EDV99075.1 QOIP01000001 RLU27361.1 CH933807 EDW11928.1 CH902620 EDV31641.1 GDIP01081470 JAM22245.1 JXJN01026397 JXJN01026508 KK107260 EZA54150.1 CP012523 ALC39997.1 CH981752 CM002910 EDX15386.1 KMY91131.1 AE014134 AY071439 KX531312 AAF53875.1 AAL49061.1 ANY27122.1 CH954179 EDV54579.1 CM000158 EDW90494.2 CH940654 EDW57899.1 CH480822 EDW55842.1 CH964214 EDW80409.1 LJIG01000253 KRT86687.1 KK122154 KFM82054.1
JAA77894.1 NWSH01007107 PCG63090.1 PCG63089.1 GDQN01008728 GDQN01007990 JAT82326.1 JAT83064.1 RSAL01000257 RVE43411.1 ODYU01004478 SOQ44386.1 GDQN01003239 JAT87815.1 PCG63088.1 JTDY01007558 KOB65114.1 AK405418 BAM20736.1 GDAI01001713 JAI15890.1 KQ459606 KPI91357.1 KQ459896 KPJ19469.1 AGBW02012813 OWR44406.1 KK852898 KDR14167.1 NNAY01000669 OXU27084.1 KC571943 AGM32442.1 PYGN01004758 PYGN01000725 PSN29271.1 PSN41633.1 GBYB01007959 JAG77726.1 DS231854 EDS38059.1 AJWK01001675 GBYB01007960 JAG77727.1 GANO01000393 JAB59478.1 CH477232 EAT46877.1 GAPW01005841 GAPW01005840 JAC07757.1 GDHF01021284 JAI31030.1 AJVK01006816 CCAG010014651 AXCM01001558 KQ982893 KYQ49595.1 GAKP01023419 JAC35539.1 GAMD01002508 JAA99082.1 GGFK01011989 MBW45310.1 GGFJ01010553 MBW59694.1 GL888528 EGI59749.1 ADTU01014508 GEDC01011023 JAS26275.1 ATLV01014737 KE524984 KFB39482.1 GAMC01007656 GAMC01007654 JAB98901.1 KQ976514 KYM82280.1 KQ981494 KYN40933.1 KQ977279 KYN04389.1 GL452471 EFN77161.1 GFDF01008319 JAV05765.1 GDIP01246519 JAI76882.1 GFDG01001969 JAV16830.1 GGFJ01010829 MBW59970.1 GFDF01008318 JAV05766.1 GDIP01129461 JAL74253.1 KQ414605 KOC69636.1 GFDG01002774 JAV16025.1 LBMM01000784 KMQ97508.1 GL732551 EFX79482.1 GL442767 EFN63074.1 ADMH02000877 ETN64792.1 KZ288376 PBC26665.1 GEZM01015027 JAV91920.1 DS235881 EEB20018.1 AAAB01008964 EAA12355.3 JRES01000914 KNC27281.1 LRGB01000944 KZS14579.1 CH379061 EAL33163.1 CH479189 EDW25573.1 JXUM01017001 KQ560472 KXJ82233.1 GDIP01248200 JAI75201.1 OUUW01000010 SPP85977.1 CH916372 EDV99075.1 QOIP01000001 RLU27361.1 CH933807 EDW11928.1 CH902620 EDV31641.1 GDIP01081470 JAM22245.1 JXJN01026397 JXJN01026508 KK107260 EZA54150.1 CP012523 ALC39997.1 CH981752 CM002910 EDX15386.1 KMY91131.1 AE014134 AY071439 KX531312 AAF53875.1 AAL49061.1 ANY27122.1 CH954179 EDV54579.1 CM000158 EDW90494.2 CH940654 EDW57899.1 CH480822 EDW55842.1 CH964214 EDW80409.1 LJIG01000253 KRT86687.1 KK122154 KFM82054.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000053268
UP000053240
+ More
UP000007151 UP000027135 UP000215335 UP000002358 UP000245037 UP000002320 UP000092461 UP000092445 UP000008820 UP000075900 UP000076408 UP000092462 UP000092444 UP000078200 UP000075883 UP000075901 UP000075809 UP000007755 UP000005205 UP000030765 UP000078540 UP000078541 UP000078542 UP000008237 UP000095301 UP000095300 UP000053825 UP000036403 UP000000305 UP000192221 UP000000311 UP000000673 UP000242457 UP000005203 UP000009046 UP000075902 UP000007062 UP000091820 UP000037069 UP000076858 UP000001819 UP000008744 UP000092443 UP000069940 UP000249989 UP000268350 UP000001070 UP000279307 UP000009192 UP000007801 UP000092460 UP000053097 UP000092553 UP000000304 UP000000803 UP000008711 UP000002282 UP000008792 UP000001292 UP000007798 UP000054359
UP000007151 UP000027135 UP000215335 UP000002358 UP000245037 UP000002320 UP000092461 UP000092445 UP000008820 UP000075900 UP000076408 UP000092462 UP000092444 UP000078200 UP000075883 UP000075901 UP000075809 UP000007755 UP000005205 UP000030765 UP000078540 UP000078541 UP000078542 UP000008237 UP000095301 UP000095300 UP000053825 UP000036403 UP000000305 UP000192221 UP000000311 UP000000673 UP000242457 UP000005203 UP000009046 UP000075902 UP000007062 UP000091820 UP000037069 UP000076858 UP000001819 UP000008744 UP000092443 UP000069940 UP000249989 UP000268350 UP000001070 UP000279307 UP000009192 UP000007801 UP000092460 UP000053097 UP000092553 UP000000304 UP000000803 UP000008711 UP000002282 UP000008792 UP000001292 UP000007798 UP000054359
Pfam
PF02269 TFIID-18kDa
SUPFAM
SSF47113
SSF47113
Gene 3D
CDD
ProteinModelPortal
Q1HQ71
H9JSE8
Q1HQ72
S4NLJ7
A0A2A4IVC1
A0A2A4ITT2
+ More
A0A1E1W7Y8 A0A3S2NBV3 A0A2H1VUF6 A0A1E1WLG1 A0A2A4IUL8 A0A0L7KQ66 I4DS46 A0A0K8TPR7 A0A194PDM5 A0A194RPY7 A0A212ESD1 A0A067QVU2 A0A232F8B8 K7IR23 R4V1D4 A0A2P8XB90 A0A0C9QW43 B0W7M4 A0A1B0C904 A0A1B0AH71 A0A0C9RI76 U5ENY6 Q17JG3 A0A023EEG2 A0A0K8UXD3 A0A182RC81 A0A182Y020 A0A1B0DM14 A0A1B0G2F6 A0A1A9UD09 A0A182MBD1 A0A182STT5 A0A151WPH5 A0A034UZ26 T1E8S3 A0A2M4AX09 A0A2M4C319 F4X177 A0A158NFQ2 A0A1B6DKU2 A0A084VND8 W8BZX5 A0A195BDM0 A0A151JY37 A0A195CW55 E2C4H7 A0A1L8DHA1 A0A0P4XBQ4 A0A1L8EDN2 A0A2M4C3R6 A0A1L8DH42 A0A1I8M6K8 A0A1I8QB52 A0A0P5TCC7 A0A0L7RFX2 A0A1L8EBM8 A0A0J7L4M6 E9GLB8 A0A1W4VXW9 E2AU31 W5JK90 A0A2A3E4N3 A0A087ZX98 A0A1Y1N4V2 E0W312 A0A182TSS8 Q7Q3U1 A0A1A9WHG0 A0A0L0C4W3 A0A164XUE7 Q29KL1 B4GT64 A0A1A9YRT3 A0A182GSK9 A0A0P4X1R9 A0A3B0JV47 B4JQ95 A0A3L8E3T7 B4KEV9 B3MKW0 A0A0P5WUM4 A0A1B0AKN3 A0A1B0C698 A0A026WDJ0 A0A0M4EDP2 B4NS08 Q9VIP1 B3NL01 B4PBI0 B4M9M3 B4I636 B4N7S0 A0A0T6BHD0 A0A087UXG5
A0A1E1W7Y8 A0A3S2NBV3 A0A2H1VUF6 A0A1E1WLG1 A0A2A4IUL8 A0A0L7KQ66 I4DS46 A0A0K8TPR7 A0A194PDM5 A0A194RPY7 A0A212ESD1 A0A067QVU2 A0A232F8B8 K7IR23 R4V1D4 A0A2P8XB90 A0A0C9QW43 B0W7M4 A0A1B0C904 A0A1B0AH71 A0A0C9RI76 U5ENY6 Q17JG3 A0A023EEG2 A0A0K8UXD3 A0A182RC81 A0A182Y020 A0A1B0DM14 A0A1B0G2F6 A0A1A9UD09 A0A182MBD1 A0A182STT5 A0A151WPH5 A0A034UZ26 T1E8S3 A0A2M4AX09 A0A2M4C319 F4X177 A0A158NFQ2 A0A1B6DKU2 A0A084VND8 W8BZX5 A0A195BDM0 A0A151JY37 A0A195CW55 E2C4H7 A0A1L8DHA1 A0A0P4XBQ4 A0A1L8EDN2 A0A2M4C3R6 A0A1L8DH42 A0A1I8M6K8 A0A1I8QB52 A0A0P5TCC7 A0A0L7RFX2 A0A1L8EBM8 A0A0J7L4M6 E9GLB8 A0A1W4VXW9 E2AU31 W5JK90 A0A2A3E4N3 A0A087ZX98 A0A1Y1N4V2 E0W312 A0A182TSS8 Q7Q3U1 A0A1A9WHG0 A0A0L0C4W3 A0A164XUE7 Q29KL1 B4GT64 A0A1A9YRT3 A0A182GSK9 A0A0P4X1R9 A0A3B0JV47 B4JQ95 A0A3L8E3T7 B4KEV9 B3MKW0 A0A0P5WUM4 A0A1B0AKN3 A0A1B0C698 A0A026WDJ0 A0A0M4EDP2 B4NS08 Q9VIP1 B3NL01 B4PBI0 B4M9M3 B4I636 B4N7S0 A0A0T6BHD0 A0A087UXG5
PDB
6MZL
E-value=9.62032e-40,
Score=404
Ontologies
GO
PANTHER
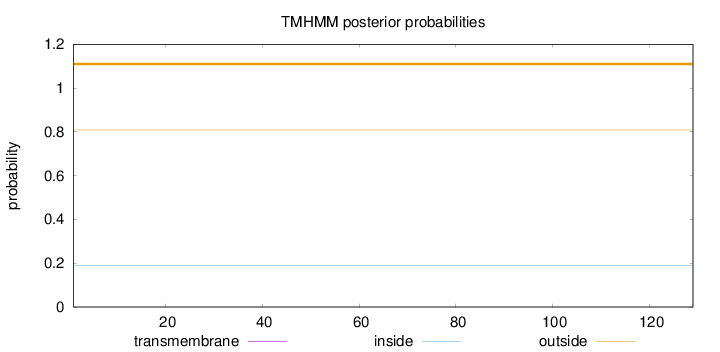
Topology
Length:
129
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.19114
outside
1 - 129
Population Genetic Test Statistics
Pi
210.435274
Theta
186.243256
Tajima's D
0.671088
CLR
0.289526
CSRT
0.564621768911554
Interpretation
Uncertain
