Gene
KWMTBOMO05105
Pre Gene Modal
BGIBMGA012511
Annotation
PREDICTED:_arylsulfatase_B-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.269
Sequence
CDS
ATGTATAACACCATTCTATTCTTGATACTGAATTGGATAGTTAACGCTTCGACTCAACAAAAATCTCCTAATATAGTTCTTATCATTGCCGATGATTTGGGATGGGACGACGTAAGTTTTCATGGATCAGACCAAATATTGACTCCGAACATCGATCTTTTGGCATACTCTGGAAAGGCCTTGGGACGCTACTATACTCACTGTATTTGTACACCATCGAGGGCAGCTCTCTTGACCGGAAAATATGCACATCGAATCGGAATGCAGGGATACCCCTTAACGAATAGTGAAGACAGAGGTTTGCCGACGACGGAAAAAATATTGCCACAATATTTGAAAGAGTTGGGATACTCGACTCATTTGGTTGGAAAGTGGCATGTTGGACAGTCACGAACTGAATATTTACCAATGCAGAGAGGTTTCGACAGTCACTTCGGACACAGAGGTGGATATGTAGACTACTATGAGTACACATTAGACGAAACGTGGAGTACGGGAGAGGTTACAGGATTTGGATTATTTCGAAATGAAACACCTGCTTGGGACGTTGAAGGTTACCTTACAGATGTTTATACTGAAGAAGCCAAATCTGTTATCAAACTTCATGATGTAAATAAGCCGCTATTTTTGATGTTAGCCCATAACGCGCCTCACGCTGCAAATTATCCTTCCATTGTACAAGCGCCACCTGAAGACATTCGAAGTATGAAACACGTGGAATTAACGGGGCGACGAACTTATGCAGCCATGGTAAAAAAGCTGGATGAAAGTGTAGGGGAAATAGTAGAGGCTTTGTATAAGAAACAGATGCTTGAAAATACAGTTATCGTGTTCATTGCTGACAATGGCGGTATGACCTCTGGGGTTGCTATGAACTTTGCATCTAATTGGCCTCTGCGAGGACTGAAAATGTCGCCATTCGAGGGAGGCGTACGTGCCACAGGACTTGTATGGAGTTCGAGTTTTAATAATTCAAACCACTTGTGGAATGGTTTTATCCACGTGACAGATTGGCTACCAATTTTTTTGAGAATAGCAGGCTCAGAAGTACCTCTTGATATTAATGGCGTAGATCTTTGGGATGACATAAACACAAAGGATAATTCCAGGCGTGATGAAATTTATGAAATTGACGACTACACCGGTTTTGCTGCTATTATATCTCAAGATTTCAAATTAGTTTCTGGACAAGTGACATTAGAATACAGCAATTATCAGGGAGAAAACCTCAGAGGGATGATAGGGGACCCACCATCTTATGCTGATTCTGTCCAGAAGAGCAAAACGTATCGTATTCTTAAAAATATTGGGAAACCTTTTAAAAAAGAGGATGTAAACCTACGTAGAAACATTAGAGTGAAATGTAATCAAGAAATAGATAAAATATGCTATCCCGATAAAAATAAGTTTTGTTTGTATAATATTAAAGAAGATCCTTGCGAAACAAACGATCTGTCCGCGACATATCCAGATATAGCGGAAAGTATGTTGAAACGTTTACATATTGAAATATCTAAAACAATACCTAGAACAGTGCCCTTGTACAGAAATCCGGAATCTTTACCTCGTCTACACAACTTCACTTGGACAACATTTGCAGACAATTTCAAATAG
Protein
MYNTILFLILNWIVNASTQQKSPNIVLIIADDLGWDDVSFHGSDQILTPNIDLLAYSGKALGRYYTHCICTPSRAALLTGKYAHRIGMQGYPLTNSEDRGLPTTEKILPQYLKELGYSTHLVGKWHVGQSRTEYLPMQRGFDSHFGHRGGYVDYYEYTLDETWSTGEVTGFGLFRNETPAWDVEGYLTDVYTEEAKSVIKLHDVNKPLFLMLAHNAPHAANYPSIVQAPPEDIRSMKHVELTGRRTYAAMVKKLDESVGEIVEALYKKQMLENTVIVFIADNGGMTSGVAMNFASNWPLRGLKMSPFEGGVRATGLVWSSSFNNSNHLWNGFIHVTDWLPIFLRIAGSEVPLDINGVDLWDDINTKDNSRRDEIYEIDDYTGFAAIISQDFKLVSGQVTLEYSNYQGENLRGMIGDPPSYADSVQKSKTYRILKNIGKPFKKEDVNLRRNIRVKCNQEIDKICYPDKNKFCLYNIKEDPCETNDLSATYPDIAESMLKRLHIEISKTIPRTVPLYRNPESLPRLHNFTWTTFADNFK
Summary
Uniprot
H9JSK0
A0A194PE98
A0A194RNQ9
A0A2H1W2J1
A0A3G5BLY0
A0A3G5BLS3
+ More
A0A3G5BLX5 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 Q8MM72 Q8MPI1 A0A3G5BLU7 A0A3G5BLV1 A0A3G5BLV4 A0A194RPN9 A0A3G5BM44 A0A194PF78 A0A194RQ78 A0A3G5BLV9 A0A3G5BLS9 A0A3G5BLW6 A0A3G5BLU4 A0A2J7QEX8 A0A336MDU4 A0A336MIA9 A0A336MLP5 A0A1B6DH44 A0A2J7RFU7 A0A3L8D7K7 B0WV91 A0A067R9K3 A0A1Q3FA56 D2A034 A0A2H1WSG7 A0A212ETP0 E0W490 A0A158P1M3 J9KB05 T1I2L4 A0A151WFD5 D2A3E0 A0A2J7PFL2 A0A067R8L8 E2AZK6 A0A2J7PFK0 A0A2J7PFJ8 A0A0T6B214 A0A088AUJ3 K7IR19 A0A1J1IM47 A0A1L8E4E2 A0A1B6MKB7 A0A2A4JRQ3 A0A3G5BLV6 A0A0K8S8T4 A0A1A9WWP8 A0A067RI69 Q5TMW7 A0A1I8MCF7 A0A0K8S7A3 A0A1B6BZR2 A0A0C9R819 A0A182YCD3 A0A182KTE9 A0A182VGM7 W5JGQ0 A0A2S2QT45 A0A182X7T1 A0A2J7RFV4 A0A067RK57 A0A1B6G4I4 A0A067QYG0 A0A182NPR4 A0A2S2R7V2 A0A336MNQ6 D2A5N6 A0A1Y1N921 A0A2H8TRW6 A0A067QXZ3 A0A195BRC3 A0A182HHX8 A0A067QUM9 A0A2S2P2T4 A0A1L8E4Y0 A0A1W4X523 A0A0L7RJP2 A0A154PHM0 E0W496 A0A1W4VDC2 A0A182RSS7 A0A087ZZX8 A0A2P8Y6M3 A0A084VIL9 A0A026W1J0 A0A0K8U1B6 A0A0K8UUD7 A0A384WE16 A0A182PV62
A0A3G5BLX5 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 Q8MM72 Q8MPI1 A0A3G5BLU7 A0A3G5BLV1 A0A3G5BLV4 A0A194RPN9 A0A3G5BM44 A0A194PF78 A0A194RQ78 A0A3G5BLV9 A0A3G5BLS9 A0A3G5BLW6 A0A3G5BLU4 A0A2J7QEX8 A0A336MDU4 A0A336MIA9 A0A336MLP5 A0A1B6DH44 A0A2J7RFU7 A0A3L8D7K7 B0WV91 A0A067R9K3 A0A1Q3FA56 D2A034 A0A2H1WSG7 A0A212ETP0 E0W490 A0A158P1M3 J9KB05 T1I2L4 A0A151WFD5 D2A3E0 A0A2J7PFL2 A0A067R8L8 E2AZK6 A0A2J7PFK0 A0A2J7PFJ8 A0A0T6B214 A0A088AUJ3 K7IR19 A0A1J1IM47 A0A1L8E4E2 A0A1B6MKB7 A0A2A4JRQ3 A0A3G5BLV6 A0A0K8S8T4 A0A1A9WWP8 A0A067RI69 Q5TMW7 A0A1I8MCF7 A0A0K8S7A3 A0A1B6BZR2 A0A0C9R819 A0A182YCD3 A0A182KTE9 A0A182VGM7 W5JGQ0 A0A2S2QT45 A0A182X7T1 A0A2J7RFV4 A0A067RK57 A0A1B6G4I4 A0A067QYG0 A0A182NPR4 A0A2S2R7V2 A0A336MNQ6 D2A5N6 A0A1Y1N921 A0A2H8TRW6 A0A067QXZ3 A0A195BRC3 A0A182HHX8 A0A067QUM9 A0A2S2P2T4 A0A1L8E4Y0 A0A1W4X523 A0A0L7RJP2 A0A154PHM0 E0W496 A0A1W4VDC2 A0A182RSS7 A0A087ZZX8 A0A2P8Y6M3 A0A084VIL9 A0A026W1J0 A0A0K8U1B6 A0A0K8UUD7 A0A384WE16 A0A182PV62
Pubmed
EMBL
BABH01027988
KQ459606
KPI91353.1
KQ459896
KPJ19473.1
ODYU01005911
+ More
SOQ47291.1 MG022102 AYW00831.1 MG022101 AYW00830.1 MG022103 AYW00832.1 MG022099 AYW00828.1 MG022100 AYW00829.1 MG022096 AJ489522 AYW00825.1 CAD33828.1 AJ410305 AJ410306 CAC86342.1 AJ410304 CAC86338.1 MG022098 AYW00827.1 MG022097 AYW00826.1 MG022108 AYW00837.1 KPJ19472.1 MG022105 AYW00834.1 KPI91354.1 KPJ19475.1 MG022107 AYW00836.1 MG022104 AYW00833.1 MG022109 AYW00838.1 MG022106 AYW00835.1 NEVH01015305 PNF27140.1 UFQT01001035 SSX28572.1 SSX28573.1 UFQS01001659 UFQT01001659 SSX11767.1 SSX31332.1 GEDC01012740 GEDC01012295 JAS24558.1 JAS25003.1 NEVH01004410 PNF39713.1 QOIP01000012 RLU15878.1 DS232119 EDS35430.1 KK852655 KDR19289.1 GFDL01010662 JAV24383.1 KQ971338 EFA01751.1 ODYU01010728 SOQ56015.1 AGBW02012542 OWR44858.1 DS235886 EEB20446.1 ADTU01001092 ABLF02026482 ACPB03016276 ACPB03016277 KQ983214 KYQ46561.1 EFA02301.1 NEVH01025657 PNF15107.1 KK852631 KDR19827.1 GL444248 EFN61139.1 PNF15112.1 PNF15110.1 LJIG01016161 KRT81432.1 CVRI01000054 CRL00636.1 GFDF01000493 JAV13591.1 GEBQ01003560 JAT36417.1 NWSH01000745 PCG74446.1 MG022110 AYW00839.1 GBRD01016667 GDHC01012310 JAG49160.1 JAQ06319.1 KK852665 KDR18961.1 AAAB01008986 EAL38777.3 GBRD01016664 GDHC01015135 JAG49163.1 JAQ03494.1 GEDC01030550 JAS06748.1 GBYB01004180 JAG73947.1 ADMH02001585 ETN61974.1 GGMS01011701 MBY80904.1 PNF39716.1 KDR19829.1 GECZ01012437 JAS57332.1 KK852878 KDR14503.1 GGMS01016567 MBY85770.1 SSX11770.1 SSX31335.1 KQ971345 EFA05395.2 GEZM01009631 JAV94444.1 GFXV01005138 MBW16943.1 KK852842 KDR15167.1 KQ976418 KYM89484.1 APCN01002450 KK852917 KDR13867.1 GGMR01011065 MBY23684.1 GFDF01000492 JAV13592.1 KQ414579 KOC71090.1 KQ434910 KZC11351.1 EEB20452.1 PYGN01000874 PSN39814.1 ATLV01013369 KE524854 KFB37813.1 KK107488 EZA49888.1 GDHF01032011 JAI20303.1 GDHF01022113 JAI30201.1 KX986117 ATE50179.1
SOQ47291.1 MG022102 AYW00831.1 MG022101 AYW00830.1 MG022103 AYW00832.1 MG022099 AYW00828.1 MG022100 AYW00829.1 MG022096 AJ489522 AYW00825.1 CAD33828.1 AJ410305 AJ410306 CAC86342.1 AJ410304 CAC86338.1 MG022098 AYW00827.1 MG022097 AYW00826.1 MG022108 AYW00837.1 KPJ19472.1 MG022105 AYW00834.1 KPI91354.1 KPJ19475.1 MG022107 AYW00836.1 MG022104 AYW00833.1 MG022109 AYW00838.1 MG022106 AYW00835.1 NEVH01015305 PNF27140.1 UFQT01001035 SSX28572.1 SSX28573.1 UFQS01001659 UFQT01001659 SSX11767.1 SSX31332.1 GEDC01012740 GEDC01012295 JAS24558.1 JAS25003.1 NEVH01004410 PNF39713.1 QOIP01000012 RLU15878.1 DS232119 EDS35430.1 KK852655 KDR19289.1 GFDL01010662 JAV24383.1 KQ971338 EFA01751.1 ODYU01010728 SOQ56015.1 AGBW02012542 OWR44858.1 DS235886 EEB20446.1 ADTU01001092 ABLF02026482 ACPB03016276 ACPB03016277 KQ983214 KYQ46561.1 EFA02301.1 NEVH01025657 PNF15107.1 KK852631 KDR19827.1 GL444248 EFN61139.1 PNF15112.1 PNF15110.1 LJIG01016161 KRT81432.1 CVRI01000054 CRL00636.1 GFDF01000493 JAV13591.1 GEBQ01003560 JAT36417.1 NWSH01000745 PCG74446.1 MG022110 AYW00839.1 GBRD01016667 GDHC01012310 JAG49160.1 JAQ06319.1 KK852665 KDR18961.1 AAAB01008986 EAL38777.3 GBRD01016664 GDHC01015135 JAG49163.1 JAQ03494.1 GEDC01030550 JAS06748.1 GBYB01004180 JAG73947.1 ADMH02001585 ETN61974.1 GGMS01011701 MBY80904.1 PNF39716.1 KDR19829.1 GECZ01012437 JAS57332.1 KK852878 KDR14503.1 GGMS01016567 MBY85770.1 SSX11770.1 SSX31335.1 KQ971345 EFA05395.2 GEZM01009631 JAV94444.1 GFXV01005138 MBW16943.1 KK852842 KDR15167.1 KQ976418 KYM89484.1 APCN01002450 KK852917 KDR13867.1 GGMR01011065 MBY23684.1 GFDF01000492 JAV13592.1 KQ414579 KOC71090.1 KQ434910 KZC11351.1 EEB20452.1 PYGN01000874 PSN39814.1 ATLV01013369 KE524854 KFB37813.1 KK107488 EZA49888.1 GDHF01032011 JAI20303.1 GDHF01022113 JAI30201.1 KX986117 ATE50179.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000235965
UP000279307
UP000002320
+ More
UP000027135 UP000007266 UP000007151 UP000009046 UP000005205 UP000007819 UP000015103 UP000075809 UP000000311 UP000005203 UP000002358 UP000183832 UP000218220 UP000091820 UP000007062 UP000095301 UP000076408 UP000075882 UP000075903 UP000000673 UP000076407 UP000075884 UP000078540 UP000075840 UP000192223 UP000053825 UP000076502 UP000192221 UP000075900 UP000245037 UP000030765 UP000053097 UP000075885
UP000027135 UP000007266 UP000007151 UP000009046 UP000005205 UP000007819 UP000015103 UP000075809 UP000000311 UP000005203 UP000002358 UP000183832 UP000218220 UP000091820 UP000007062 UP000095301 UP000076408 UP000075882 UP000075903 UP000000673 UP000076407 UP000075884 UP000078540 UP000075840 UP000192223 UP000053825 UP000076502 UP000192221 UP000075900 UP000245037 UP000030765 UP000053097 UP000075885
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JSK0
A0A194PE98
A0A194RNQ9
A0A2H1W2J1
A0A3G5BLY0
A0A3G5BLS3
+ More
A0A3G5BLX5 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 Q8MM72 Q8MPI1 A0A3G5BLU7 A0A3G5BLV1 A0A3G5BLV4 A0A194RPN9 A0A3G5BM44 A0A194PF78 A0A194RQ78 A0A3G5BLV9 A0A3G5BLS9 A0A3G5BLW6 A0A3G5BLU4 A0A2J7QEX8 A0A336MDU4 A0A336MIA9 A0A336MLP5 A0A1B6DH44 A0A2J7RFU7 A0A3L8D7K7 B0WV91 A0A067R9K3 A0A1Q3FA56 D2A034 A0A2H1WSG7 A0A212ETP0 E0W490 A0A158P1M3 J9KB05 T1I2L4 A0A151WFD5 D2A3E0 A0A2J7PFL2 A0A067R8L8 E2AZK6 A0A2J7PFK0 A0A2J7PFJ8 A0A0T6B214 A0A088AUJ3 K7IR19 A0A1J1IM47 A0A1L8E4E2 A0A1B6MKB7 A0A2A4JRQ3 A0A3G5BLV6 A0A0K8S8T4 A0A1A9WWP8 A0A067RI69 Q5TMW7 A0A1I8MCF7 A0A0K8S7A3 A0A1B6BZR2 A0A0C9R819 A0A182YCD3 A0A182KTE9 A0A182VGM7 W5JGQ0 A0A2S2QT45 A0A182X7T1 A0A2J7RFV4 A0A067RK57 A0A1B6G4I4 A0A067QYG0 A0A182NPR4 A0A2S2R7V2 A0A336MNQ6 D2A5N6 A0A1Y1N921 A0A2H8TRW6 A0A067QXZ3 A0A195BRC3 A0A182HHX8 A0A067QUM9 A0A2S2P2T4 A0A1L8E4Y0 A0A1W4X523 A0A0L7RJP2 A0A154PHM0 E0W496 A0A1W4VDC2 A0A182RSS7 A0A087ZZX8 A0A2P8Y6M3 A0A084VIL9 A0A026W1J0 A0A0K8U1B6 A0A0K8UUD7 A0A384WE16 A0A182PV62
A0A3G5BLX5 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 Q8MM72 Q8MPI1 A0A3G5BLU7 A0A3G5BLV1 A0A3G5BLV4 A0A194RPN9 A0A3G5BM44 A0A194PF78 A0A194RQ78 A0A3G5BLV9 A0A3G5BLS9 A0A3G5BLW6 A0A3G5BLU4 A0A2J7QEX8 A0A336MDU4 A0A336MIA9 A0A336MLP5 A0A1B6DH44 A0A2J7RFU7 A0A3L8D7K7 B0WV91 A0A067R9K3 A0A1Q3FA56 D2A034 A0A2H1WSG7 A0A212ETP0 E0W490 A0A158P1M3 J9KB05 T1I2L4 A0A151WFD5 D2A3E0 A0A2J7PFL2 A0A067R8L8 E2AZK6 A0A2J7PFK0 A0A2J7PFJ8 A0A0T6B214 A0A088AUJ3 K7IR19 A0A1J1IM47 A0A1L8E4E2 A0A1B6MKB7 A0A2A4JRQ3 A0A3G5BLV6 A0A0K8S8T4 A0A1A9WWP8 A0A067RI69 Q5TMW7 A0A1I8MCF7 A0A0K8S7A3 A0A1B6BZR2 A0A0C9R819 A0A182YCD3 A0A182KTE9 A0A182VGM7 W5JGQ0 A0A2S2QT45 A0A182X7T1 A0A2J7RFV4 A0A067RK57 A0A1B6G4I4 A0A067QYG0 A0A182NPR4 A0A2S2R7V2 A0A336MNQ6 D2A5N6 A0A1Y1N921 A0A2H8TRW6 A0A067QXZ3 A0A195BRC3 A0A182HHX8 A0A067QUM9 A0A2S2P2T4 A0A1L8E4Y0 A0A1W4X523 A0A0L7RJP2 A0A154PHM0 E0W496 A0A1W4VDC2 A0A182RSS7 A0A087ZZX8 A0A2P8Y6M3 A0A084VIL9 A0A026W1J0 A0A0K8U1B6 A0A0K8UUD7 A0A384WE16 A0A182PV62
PDB
1FSU
E-value=6.98678e-65,
Score=629
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.982055
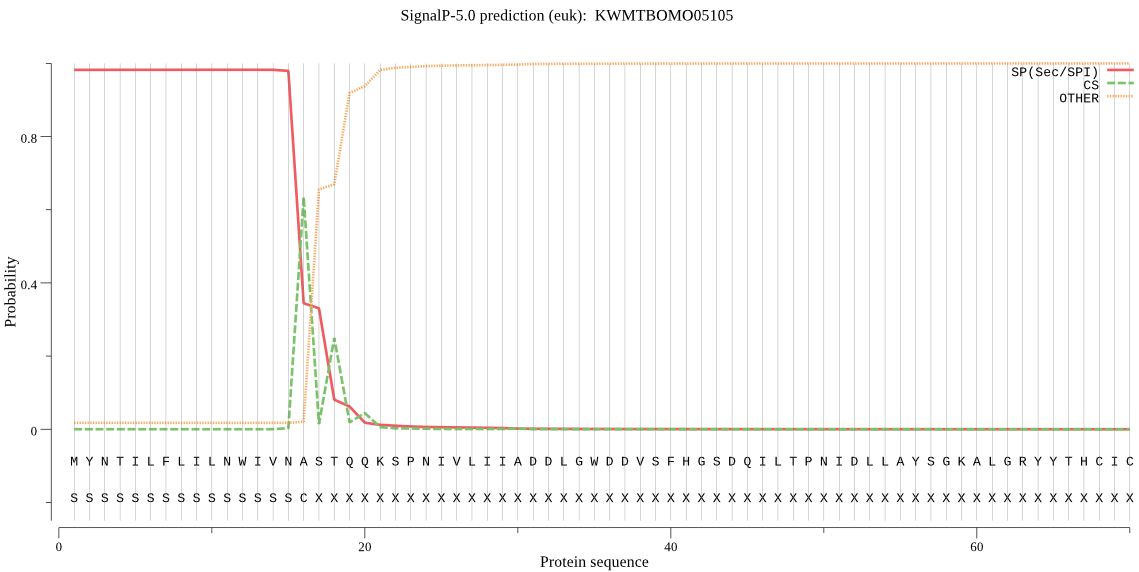
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0862600000000001
Exp number, first 60 AAs:
0.00773
Total prob of N-in:
0.00295
outside
1 - 537
Population Genetic Test Statistics
Pi
238.697086
Theta
191.181361
Tajima's D
1.298623
CLR
0.000265
CSRT
0.740162991850407
Interpretation
Uncertain
