Gene
KWMTBOMO05104
Pre Gene Modal
BGIBMGA012457
Annotation
_odorant_receptor_30_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.071
Sequence
CDS
ATGTCGGTTTCCAATTTGAAATTCGAAGTACTTTTCAAACCTACCACTATGTCATTGCATATGAATCGTTCGCATCCATCTATCAAACGAAATAAAATTTGGCTACTCCAGTTCATATCACTAATGACGCTTACGGTATTTTGTGCTACTGGACTAATCACATCCTTATTGTTCCACGATTTAAAATTTGGTAAATACATGGAAGCAAGCAAGAATGGAACTATAGCTATGCTGTCATTTACAACCACTTTTAAATACAGTCTTTTGCTTTATTTACAAAAATCTCTCAACCGTCTCATTGCAAAAATTGACATGGATTACGAAATTGCAAAAGGCTTAACACCTCAGGAAAAGGCTATAGTGCTTAATTATGCTAAAAAGGGAGTCATTGTGAGCAAATTCTGGTTATTCACAGCATTTGCAATAACATTTTGTTTTCCCCTAAAGGCTTTTATTATTATGGGTTACCGATTCTTTATCAAAAACGAATTTCGGCTAGAGCCAATGTTTGACATGACATACCCAGAACCTATAGAAAGTTACAAAACTTCGTTTCCTGTTTACTTTATTCTATTTGTCGTGTTTTTTTTGTTTGGTTGTTATGCTTCAAGTTTGTTTGTGGCATTTGATCCTCTAGTGCCTATCTTCGTGTTACATGCTTGTGGACAACTTGATTTACTTAGCCTAAGAATCACCAAACTGTTTTCGGACACAAAAAACCCGAGAATTATTGCCAAGGAATTGAAAGTGATAATATCCAAGCTGCAGGAGCTGTACGGCTTTGTGAATTTCATCAAGGTCAACTTTTCTATTTTATATGAATATAATATGAAAATTACCACAATCTCAATGCCACTAAGTGCTTTTCAAGTTGTAGAGAGTCTGCGGCGGGGAGAATTTAATATAGAGTTTACATACTTCTTTTTTGGATGTATACTTCACTTTTTTATGCCATGCTATTACAGCAATCTTCTTATGGAAAGGAGTGAGAATTTTCGATTCGCCATATATTCATGCGGTTGGGAAAACCATAATGATAAAAATATTCGACAAATGCTGTTGTTCATGTTAACCAGAGCCACAGAACCGCTCGGTATAGCTACCGTATTTACCAACATTTCTCTCGACACATTTGCCGAGGTTAATACTTTTGACACGGTTTTATTAGCTTGA
Protein
MSVSNLKFEVLFKPTTMSLHMNRSHPSIKRNKIWLLQFISLMTLTVFCATGLITSLLFHDLKFGKYMEASKNGTIAMLSFTTTFKYSLLLYLQKSLNRLIAKIDMDYEIAKGLTPQEKAIVLNYAKKGVIVSKFWLFTAFAITFCFPLKAFIIMGYRFFIKNEFRLEPMFDMTYPEPIESYKTSFPVYFILFVVFFLFGCYASSLFVAFDPLVPIFVLHACGQLDLLSLRITKLFSDTKNPRIIAKELKVIISKLQELYGFVNFIKVNFSILYEYNMKITTISMPLSAFQVVESLRRGEFNIEFTYFFFGCILHFFMPCYYSNLLMERSENFRFAIYSCGWENHNDKNIRQMLLFMLTRATEPLGIATVFTNISLDTFAEVNTFDTVLLA
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
A7E3H1
C4B7W1
Q0EEG0
A0A0P1IW45
A0A2L1TG23
A0A0S1TR03
+ More
A0A1B3B735 A0A223HD14 H9A5P8 A0A0V0J2J7 A0A345BEV6 A0A223HDE4 A0A097ITS2 A0A0B5CVK0 A0A097IYK0 A0A0B5GR49 A0A0E4B5D2 A0A0K8TUW5 A0A386H932 A0A1Y9TKA6 A0A223HD58 A0A1B3P5S5 A0A2H1X5G9 A0A3S2LTC0 A0A1S5XXN1 A0A212F883 A0A2L1TG69 A0A2L1TG63 A0A1B3B751 A0A0K8TUR7 H9A5Q1 A0A075T803 A0A345BEW3 A0A2H1W9T2 A0A2K8GL60 A0A212ET92 A0A2A4JYE1 A0A386H9E0 A0A0K8TUS4 A0A0B5CUR9 A0A097IYN4 A7E3H5 A0A097ITU4 A7E3H4 A2T870 A0A0B5GR60 A0A0S1TQ01 A0A2K8GL57 A0A076EDH2 A0A0K8TVD8 A0A0K8TU79 A0A0K8TU70 A0A1S5XXL8 A0A2L1TG75 A0A0S1TPS3 A0A2H1W8S1 A0A2A4J2M8 A0A2A4J3K1 A0A076E9A4 A0A075T2Z7 A0A345BEW4 A0A2H1W893 A0A076E9C5 A0A076E983 A0A2K8GL33 A0A076E9C1 A0A2K8GL47 A0A2K8GL51 A0A0V0J0Y6 A0A076E999 A0A223HD27 A0A2K8GL53 A0A076E7K0 A0A2K8GL55 A0A076E7M9 A0A076E616 A0A076EDF2 C4B7W3 A0A2A4JWM6 A0A076E605 A0A3G2KX42 A0A2K8GL43 A0A076E5Y8 A0A2K8GL70 A0A3G2KX44 A0A2K8GL62 A0A223HD08
A0A1B3B735 A0A223HD14 H9A5P8 A0A0V0J2J7 A0A345BEV6 A0A223HDE4 A0A097ITS2 A0A0B5CVK0 A0A097IYK0 A0A0B5GR49 A0A0E4B5D2 A0A0K8TUW5 A0A386H932 A0A1Y9TKA6 A0A223HD58 A0A1B3P5S5 A0A2H1X5G9 A0A3S2LTC0 A0A1S5XXN1 A0A212F883 A0A2L1TG69 A0A2L1TG63 A0A1B3B751 A0A0K8TUR7 H9A5Q1 A0A075T803 A0A345BEW3 A0A2H1W9T2 A0A2K8GL60 A0A212ET92 A0A2A4JYE1 A0A386H9E0 A0A0K8TUS4 A0A0B5CUR9 A0A097IYN4 A7E3H5 A0A097ITU4 A7E3H4 A2T870 A0A0B5GR60 A0A0S1TQ01 A0A2K8GL57 A0A076EDH2 A0A0K8TVD8 A0A0K8TU79 A0A0K8TU70 A0A1S5XXL8 A0A2L1TG75 A0A0S1TPS3 A0A2H1W8S1 A0A2A4J2M8 A0A2A4J3K1 A0A076E9A4 A0A075T2Z7 A0A345BEW4 A0A2H1W893 A0A076E9C5 A0A076E983 A0A2K8GL33 A0A076E9C1 A0A2K8GL47 A0A2K8GL51 A0A0V0J0Y6 A0A076E999 A0A223HD27 A0A2K8GL53 A0A076E7K0 A0A2K8GL55 A0A076E7M9 A0A076E616 A0A076EDF2 C4B7W3 A0A2A4JWM6 A0A076E605 A0A3G2KX42 A0A2K8GL43 A0A076E5Y8 A0A2K8GL70 A0A3G2KX44 A0A2K8GL62 A0A223HD08
Pubmed
EMBL
BK005936
DAA05986.1
AB472110
BAH66327.1
AB234350
BAF31190.1
+ More
LN885108 CUQ99397.1 KY707200 AVF19642.1 KR935727 ALM26219.1 KT588122 AOE48032.1 KY283606 AST36265.1 JN836698 AFC91738.2 GDKB01000048 JAP38448.1 MG816595 AXF48780.1 KY283736 AST36395.1 KM597194 AIT69871.1 KM678295 AJE25868.1 KM655530 AIT71980.1 KM892368 AJF23783.1 LC002747 BAR43495.1 GCVX01000169 JAI18061.1 MG546619 AYD42242.1 KX084503 ARO76458.1 KY283658 AST36317.1 KX655985 AOG12934.1 ODYU01013161 SOQ59854.1 RSAL01000257 RVE43406.1 KY399279 AQQ73510.1 AGBW02009791 OWR49919.1 KY707224 AVF19666.1 KY707226 AVF19668.1 KT588104 AOE48014.1 GCVX01000197 JAI18033.1 JN836701 AFC91741.2 KF768708 AIG51902.1 MG816602 AXF48787.1 ODYU01006937 SOQ49234.1 KY225506 ARO70518.1 AGBW02012639 OWR44671.1 NWSH01000442 PCG76402.1 MG546601 AYD42224.1 GCVX01000187 JAI18043.1 KM678309 AJE25882.1 KM655546 AIT71996.1 BK005940 DAA05990.1 KM597210 AIT69887.1 BK005939 DAA05989.1 DQ991154 ABK27848.1 KM892383 AJF23798.1 KR935731 ALM26223.1 KY225517 ARO70529.1 KF487697 AII01095.1 GCVX01000190 JAI18040.1 GCVX01000193 JAI18037.1 GCVX01000191 JAI18039.1 KY399258 AQQ73489.1 KY707227 AVF19669.1 KR935732 ALM26224.1 SOQ49232.1 NWSH01003434 PCG66315.1 PCG66316.1 KF487676 AII01074.1 KF768709 AIG51903.1 MG816603 AXF48788.1 SOQ49233.1 KF487700 AII01098.1 KF487650 AII01048.1 KY225490 ARO70502.1 KF487695 AII01093.1 KY225505 ARO70517.1 KY225504 ARO70516.1 GDKB01000051 JAP38445.1 KF487671 AII01069.1 KY283612 AST36271.1 KY225509 ARO70521.1 KF487669 AII01067.1 KY225508 ARO70520.1 KF487699 AII01097.1 KF487698 AII01096.1 KF487672 AII01070.1 AB472112 BAH66329.1 PCG76401.1 KF487688 AII01086.1 MH723592 AYN64395.1 KY225494 ARO70506.1 KF487668 AII01066.1 KY225512 ARO70524.1 MH723591 AYN64394.1 KY225518 ARO70530.1 KY283611 AST36270.1
LN885108 CUQ99397.1 KY707200 AVF19642.1 KR935727 ALM26219.1 KT588122 AOE48032.1 KY283606 AST36265.1 JN836698 AFC91738.2 GDKB01000048 JAP38448.1 MG816595 AXF48780.1 KY283736 AST36395.1 KM597194 AIT69871.1 KM678295 AJE25868.1 KM655530 AIT71980.1 KM892368 AJF23783.1 LC002747 BAR43495.1 GCVX01000169 JAI18061.1 MG546619 AYD42242.1 KX084503 ARO76458.1 KY283658 AST36317.1 KX655985 AOG12934.1 ODYU01013161 SOQ59854.1 RSAL01000257 RVE43406.1 KY399279 AQQ73510.1 AGBW02009791 OWR49919.1 KY707224 AVF19666.1 KY707226 AVF19668.1 KT588104 AOE48014.1 GCVX01000197 JAI18033.1 JN836701 AFC91741.2 KF768708 AIG51902.1 MG816602 AXF48787.1 ODYU01006937 SOQ49234.1 KY225506 ARO70518.1 AGBW02012639 OWR44671.1 NWSH01000442 PCG76402.1 MG546601 AYD42224.1 GCVX01000187 JAI18043.1 KM678309 AJE25882.1 KM655546 AIT71996.1 BK005940 DAA05990.1 KM597210 AIT69887.1 BK005939 DAA05989.1 DQ991154 ABK27848.1 KM892383 AJF23798.1 KR935731 ALM26223.1 KY225517 ARO70529.1 KF487697 AII01095.1 GCVX01000190 JAI18040.1 GCVX01000193 JAI18037.1 GCVX01000191 JAI18039.1 KY399258 AQQ73489.1 KY707227 AVF19669.1 KR935732 ALM26224.1 SOQ49232.1 NWSH01003434 PCG66315.1 PCG66316.1 KF487676 AII01074.1 KF768709 AIG51903.1 MG816603 AXF48788.1 SOQ49233.1 KF487700 AII01098.1 KF487650 AII01048.1 KY225490 ARO70502.1 KF487695 AII01093.1 KY225505 ARO70517.1 KY225504 ARO70516.1 GDKB01000051 JAP38445.1 KF487671 AII01069.1 KY283612 AST36271.1 KY225509 ARO70521.1 KF487669 AII01067.1 KY225508 ARO70520.1 KF487699 AII01097.1 KF487698 AII01096.1 KF487672 AII01070.1 AB472112 BAH66329.1 PCG76401.1 KF487688 AII01086.1 MH723592 AYN64395.1 KY225494 ARO70506.1 KF487668 AII01066.1 KY225512 ARO70524.1 MH723591 AYN64394.1 KY225518 ARO70530.1 KY283611 AST36270.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
A7E3H1
C4B7W1
Q0EEG0
A0A0P1IW45
A0A2L1TG23
A0A0S1TR03
+ More
A0A1B3B735 A0A223HD14 H9A5P8 A0A0V0J2J7 A0A345BEV6 A0A223HDE4 A0A097ITS2 A0A0B5CVK0 A0A097IYK0 A0A0B5GR49 A0A0E4B5D2 A0A0K8TUW5 A0A386H932 A0A1Y9TKA6 A0A223HD58 A0A1B3P5S5 A0A2H1X5G9 A0A3S2LTC0 A0A1S5XXN1 A0A212F883 A0A2L1TG69 A0A2L1TG63 A0A1B3B751 A0A0K8TUR7 H9A5Q1 A0A075T803 A0A345BEW3 A0A2H1W9T2 A0A2K8GL60 A0A212ET92 A0A2A4JYE1 A0A386H9E0 A0A0K8TUS4 A0A0B5CUR9 A0A097IYN4 A7E3H5 A0A097ITU4 A7E3H4 A2T870 A0A0B5GR60 A0A0S1TQ01 A0A2K8GL57 A0A076EDH2 A0A0K8TVD8 A0A0K8TU79 A0A0K8TU70 A0A1S5XXL8 A0A2L1TG75 A0A0S1TPS3 A0A2H1W8S1 A0A2A4J2M8 A0A2A4J3K1 A0A076E9A4 A0A075T2Z7 A0A345BEW4 A0A2H1W893 A0A076E9C5 A0A076E983 A0A2K8GL33 A0A076E9C1 A0A2K8GL47 A0A2K8GL51 A0A0V0J0Y6 A0A076E999 A0A223HD27 A0A2K8GL53 A0A076E7K0 A0A2K8GL55 A0A076E7M9 A0A076E616 A0A076EDF2 C4B7W3 A0A2A4JWM6 A0A076E605 A0A3G2KX42 A0A2K8GL43 A0A076E5Y8 A0A2K8GL70 A0A3G2KX44 A0A2K8GL62 A0A223HD08
A0A1B3B735 A0A223HD14 H9A5P8 A0A0V0J2J7 A0A345BEV6 A0A223HDE4 A0A097ITS2 A0A0B5CVK0 A0A097IYK0 A0A0B5GR49 A0A0E4B5D2 A0A0K8TUW5 A0A386H932 A0A1Y9TKA6 A0A223HD58 A0A1B3P5S5 A0A2H1X5G9 A0A3S2LTC0 A0A1S5XXN1 A0A212F883 A0A2L1TG69 A0A2L1TG63 A0A1B3B751 A0A0K8TUR7 H9A5Q1 A0A075T803 A0A345BEW3 A0A2H1W9T2 A0A2K8GL60 A0A212ET92 A0A2A4JYE1 A0A386H9E0 A0A0K8TUS4 A0A0B5CUR9 A0A097IYN4 A7E3H5 A0A097ITU4 A7E3H4 A2T870 A0A0B5GR60 A0A0S1TQ01 A0A2K8GL57 A0A076EDH2 A0A0K8TVD8 A0A0K8TU79 A0A0K8TU70 A0A1S5XXL8 A0A2L1TG75 A0A0S1TPS3 A0A2H1W8S1 A0A2A4J2M8 A0A2A4J3K1 A0A076E9A4 A0A075T2Z7 A0A345BEW4 A0A2H1W893 A0A076E9C5 A0A076E983 A0A2K8GL33 A0A076E9C1 A0A2K8GL47 A0A2K8GL51 A0A0V0J0Y6 A0A076E999 A0A223HD27 A0A2K8GL53 A0A076E7K0 A0A2K8GL55 A0A076E7M9 A0A076E616 A0A076EDF2 C4B7W3 A0A2A4JWM6 A0A076E605 A0A3G2KX42 A0A2K8GL43 A0A076E5Y8 A0A2K8GL70 A0A3G2KX44 A0A2K8GL62 A0A223HD08
Ontologies
KEGG
GO
PANTHER
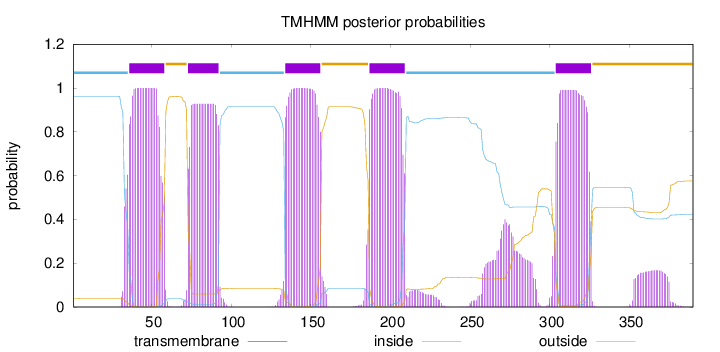
Topology
Subcellular location
Cell membrane
Length:
390
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
122.54393
Exp number, first 60 AAs:
22.84914
Total prob of N-in:
0.96083
POSSIBLE N-term signal
sequence
inside
1 - 35
TMhelix
36 - 58
outside
59 - 72
TMhelix
73 - 92
inside
93 - 133
TMhelix
134 - 156
outside
157 - 186
TMhelix
187 - 209
inside
210 - 303
TMhelix
304 - 326
outside
327 - 390
Population Genetic Test Statistics
Pi
202.075113
Theta
155.470552
Tajima's D
0.729147
CLR
0.330242
CSRT
0.580320983950802
Interpretation
Uncertain
