Pre Gene Modal
BGIBMGA012512
Annotation
PREDICTED:_arylsulfatase_B-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.126
Sequence
CDS
ATGTTTTTTACATTTTTATTTATTATTGGTACATTAATTTTAAACACGCTATCAGAAGACATAGTGAAACCTAATATAGTTTTCATTGTTGCAGATGATTTGGGATGGGACGATGTTAGCATTCACGGATCTGATCAAATCATGACACCAAATATCGACATAATGGGATATGAAGGTACAATATTGCATCAATATTACACGGATTCACTAGGGACCGCGTCGAGGAGTGCTCTTTTCACTGGAAAATACGCTATTCGTCTAGGGACTCACGGTTCATCGATAGCGGCAACAGAAGATGGTGGGGTACCGTTGTCTGAACGTCTCCTACCTTCTTATTTGCAAGATGCAGGATACACTACACATTTGCTCGGCAAATGGCAACTGGGGCATTCGAGGAAATCTTATTTGCCAACAAGCAGAGGGTTTGACACTTTCTATGGCTTCGTAACCGGATCAGTTGACTACTATACATATAGCCATGTGGAGAGTTTTAATGGATCTGCGTTTTTCGGGCTTGATTTGTTCGAAAACACGCACCCAGTTGATAATCAAACAGGTCATCTCGCTGATATTATTACAGATCAAGCTATTCAAATTATTAGAAACCATGATATAAATAAGCCGCTTTATCTCCATGTATCTCACGCTGCTCCACACGCAGGCGGTGGTTTGGTTCATTTACAAACTCCCGAAGACACAATAAACGCAAATGATCATATTGCTCACTCTGCTCGAAGATTATATGCAGGTATGGTTACAGGTCTAGATAAAAGTATTGGAAATATTATAGCAGCACTAGCAGAAAGGGAAATTCTTGATAATACATTGATTATATTTGTTTCGGACAACGGTGCTGCGCCCGTTGGGGCGACACAAAATTTCGGATCCAATTTACCCTTGAGAGGAACAAAGGGAAGACCTTGGGAGGGCGCTATTAGATCAATCGCTATAGTATGGCATGCGTCAATCACACCTCAAATCTGGAATGGATTATTCCATGTTACAGATTGGTTGCCTACTTTAGTTGCTGCAGCCGGAGGTAAAGCTCCAACTGGAATCGATGGTGTGAACCAATGGAATGCTCTTACAAAAGACGAGGACTCGAGTAGAAAGCGTTTACTATTGACAGTTGACGATTTGAACGGTTGGGCTGCGTACAGAGATGGAGATTTCAAAATAATAATCGGAGATGTTGATAAGGAAACCAGTGTTTATTTAGGAAATGAATTCAAAGAATTGCGGCTGCCAGTACATTTATACGAGAGCACGCTACTTAGTTCAGAAGCTGCTCATGTTTTTAAAGAGATTTTAGATTTATATTTGGACTTAGATGAAATATTTCATAAAAGAGATAAGCTGAGTTTACGTTCGAAACATAATATTTCACAAGAATTTAATTTTTGCGTACCATCAAGAGAATCAGGGTGTTTGTTTAACATCACAGCAGATCCTTTAGAAACCACGAATTTATGGCCTTCATTGCCTGATGTTGTGAGACGTATGGTTTTGCGACTACGTTATTACTGGGCACAAATAAACCCTAGATGTTTAGTGAAAATCGATAAAAAATCTGATCCCTCCCTTCGTGATTTTGTATGGTCACCTTGGCTAAAAGATAATGAACAAGTAGCAAAACCAATCAGAGATAGGCCACGATTTCCTCTCAAAGTTTTAGTTGGGGAATTGCAATATTTAATTGATTCAAAATTCAACAATTTTAGTGAAACAATGAGTGCCTATGTTAAATCCATGGCTGATTCGTTTTTAGAGAATGTTAGTGAATTGTTTTCGTTTTAA
Protein
MFFTFLFIIGTLILNTLSEDIVKPNIVFIVADDLGWDDVSIHGSDQIMTPNIDIMGYEGTILHQYYTDSLGTASRSALFTGKYAIRLGTHGSSIAATEDGGVPLSERLLPSYLQDAGYTTHLLGKWQLGHSRKSYLPTSRGFDTFYGFVTGSVDYYTYSHVESFNGSAFFGLDLFENTHPVDNQTGHLADIITDQAIQIIRNHDINKPLYLHVSHAAPHAGGGLVHLQTPEDTINANDHIAHSARRLYAGMVTGLDKSIGNIIAALAEREILDNTLIIFVSDNGAAPVGATQNFGSNLPLRGTKGRPWEGAIRSIAIVWHASITPQIWNGLFHVTDWLPTLVAAAGGKAPTGIDGVNQWNALTKDEDSSRKRLLLTVDDLNGWAAYRDGDFKIIIGDVDKETSVYLGNEFKELRLPVHLYESTLLSSEAAHVFKEILDLYLDLDEIFHKRDKLSLRSKHNISQEFNFCVPSRESGCLFNITADPLETTNLWPSLPDVVRRMVLRLRYYWAQINPRCLVKIDKKSDPSLRDFVWSPWLKDNEQVAKPIRDRPRFPLKVLVGELQYLIDSKFNNFSETMSAYVKSMADSFLENVSELFSF
Summary
Uniprot
H9JSK1
A0A3G5BLW6
A0A3G5BLV4
A0A194RQ78
A0A3G5BLV9
A0A3G5BM44
+ More
A0A3G5BLS9 A0A3G5BLU4 A0A194RPN9 A0A194PF78 A0A3G5BLS3 A0A3G5BLY0 A0A194PE98 A0A3G5BLX5 A0A2A4J0B7 A0A2H1W2J1 A0A194RNQ9 H9JSK0 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 A0A2H8TM77 Q8MPI1 Q8MM72 A0A3G5BLU7 A0A384WE16 A0A3Q0IUC2 D2A3E0 A0A067RK57 A0A2J7QEX8 A0A3G5BLV1 J9KB05 A0A2J7RFU7 A0A384X577 A0A2S2R7V2 A0A384WD78 A0A1Y1N921 A0A384WD96 E0W490 A0A2A3EKI1 A0A384WD97 A0A2H8TRW6 A0A1L8E4Y8 A0A2S2P2T4 A0A154PHM0 A0A0L7RJP2 A0A1B6MM89 A0A2S2QT45 A0A384WDF6 A0A1B6BZR2 A0A1L8E4N6 A0A384WXG8 A0A087ZPU2 T1HN07 A0A1L8E508 A0A384WD64 A0A1B6G4I4 A0A1B6DH44 D2A206 A0A384WT78 A0A3L8D7K7 A0A1W4XG67 A0A310SCZ1 A0A067QUM9 A0A384WT96 A0A087ZZX8 A0A336M966 A0A0A9XP28 A0A293MQ50 A0A067QYG0 A0A067R9K3 A0A0K8S8T4 A0A146M1G8 A0A2H1WSG7 A0A0K8S7A3 A0A0N0BK53 K7J2L9 A0A3G5BLV6 A0A2J7PFJ8 A0A2J7PFK0 A0A0L7QTR2 A0A158P1M3 A0A1W4X523 A0A1D2MVN0 A0A1S3DG86 A0A2P8Y6M3 A0A067R8L8 E2A1F4 A0A151WFD5 A0A3L8E386 E0W3F1 B4N5G9 T1I2L4 V3ZFY3 A0A087T2D9 A0A0A1WQR8 A0A1D1UZJ7
A0A3G5BLS9 A0A3G5BLU4 A0A194RPN9 A0A194PF78 A0A3G5BLS3 A0A3G5BLY0 A0A194PE98 A0A3G5BLX5 A0A2A4J0B7 A0A2H1W2J1 A0A194RNQ9 H9JSK0 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 A0A2H8TM77 Q8MPI1 Q8MM72 A0A3G5BLU7 A0A384WE16 A0A3Q0IUC2 D2A3E0 A0A067RK57 A0A2J7QEX8 A0A3G5BLV1 J9KB05 A0A2J7RFU7 A0A384X577 A0A2S2R7V2 A0A384WD78 A0A1Y1N921 A0A384WD96 E0W490 A0A2A3EKI1 A0A384WD97 A0A2H8TRW6 A0A1L8E4Y8 A0A2S2P2T4 A0A154PHM0 A0A0L7RJP2 A0A1B6MM89 A0A2S2QT45 A0A384WDF6 A0A1B6BZR2 A0A1L8E4N6 A0A384WXG8 A0A087ZPU2 T1HN07 A0A1L8E508 A0A384WD64 A0A1B6G4I4 A0A1B6DH44 D2A206 A0A384WT78 A0A3L8D7K7 A0A1W4XG67 A0A310SCZ1 A0A067QUM9 A0A384WT96 A0A087ZZX8 A0A336M966 A0A0A9XP28 A0A293MQ50 A0A067QYG0 A0A067R9K3 A0A0K8S8T4 A0A146M1G8 A0A2H1WSG7 A0A0K8S7A3 A0A0N0BK53 K7J2L9 A0A3G5BLV6 A0A2J7PFJ8 A0A2J7PFK0 A0A0L7QTR2 A0A158P1M3 A0A1W4X523 A0A1D2MVN0 A0A1S3DG86 A0A2P8Y6M3 A0A067R8L8 E2A1F4 A0A151WFD5 A0A3L8E386 E0W3F1 B4N5G9 T1I2L4 V3ZFY3 A0A087T2D9 A0A0A1WQR8 A0A1D1UZJ7
Pubmed
EMBL
BABH01027997
MG022109
AYW00838.1
MG022108
AYW00837.1
KQ459896
+ More
KPJ19475.1 MG022107 AYW00836.1 MG022105 AYW00834.1 MG022104 AYW00833.1 MG022106 AYW00835.1 KPJ19472.1 KQ459606 KPI91354.1 MG022101 AYW00830.1 MG022102 AYW00831.1 KPI91353.1 MG022103 AYW00832.1 NWSH01004439 PCG65118.1 ODYU01005911 SOQ47291.1 KPJ19473.1 BABH01027988 MG022099 AYW00828.1 MG022100 AYW00829.1 MG022096 AJ489522 AYW00825.1 CAD33828.1 GFXV01003236 MBW15041.1 AJ410304 CAC86338.1 AJ410305 AJ410306 CAC86342.1 MG022098 AYW00827.1 KX986117 ATE50179.1 KQ971338 EFA02301.1 KK852631 KDR19829.1 NEVH01015305 PNF27140.1 MG022097 AYW00826.1 ABLF02026482 NEVH01004410 PNF39713.1 KX986126 ATE50188.1 GGMS01016567 MBY85770.1 KX986120 ATE50182.1 GEZM01009631 JAV94444.1 KX986122 ATE50184.1 DS235886 EEB20446.1 KZ288227 PBC31752.1 KX986130 ATE50192.1 GFXV01005138 MBW16943.1 GFDF01000380 JAV13704.1 GGMR01011065 MBY23684.1 KQ434910 KZC11351.1 KQ414579 KOC71090.1 GEBQ01002945 JAT37032.1 GGMS01011701 MBY80904.1 KX986121 ATE50183.1 GEDC01030550 JAS06748.1 GFDF01000384 JAV13700.1 KX986124 ATE50186.1 ACPB03014568 ACPB03014569 GFDF01000385 JAV13699.1 KX986115 ATE50177.1 GECZ01012437 JAS57332.1 GEDC01012740 GEDC01012295 JAS24558.1 JAS25003.1 EFA02715.1 KX986118 ATE50180.1 QOIP01000012 RLU15878.1 KQ763209 OAD55144.1 KK852917 KDR13867.1 KX986128 ATE50190.1 UFQS01000723 UFQT01000723 SSX06412.1 SSX26765.1 GBHO01022208 JAG21396.1 GFWV01018204 MAA42932.1 KK852878 KDR14503.1 KK852655 KDR19289.1 GBRD01016667 GDHC01012310 JAG49160.1 JAQ06319.1 GDHC01005360 JAQ13269.1 ODYU01010728 SOQ56015.1 GBRD01016664 GDHC01015135 JAG49163.1 JAQ03494.1 KQ435708 KOX80112.1 MG022110 AYW00839.1 NEVH01025657 PNF15110.1 PNF15112.1 KQ414740 KOC62043.1 ADTU01001092 LJIJ01000470 ODM97123.1 PYGN01000874 PSN39814.1 KDR19827.1 GL435766 EFN72627.1 KQ983214 KYQ46561.1 QOIP01000001 RLU26669.1 DS235882 EEB20157.1 CH964101 EDW79608.1 ACPB03016276 ACPB03016277 KB202481 ESO90113.1 KK113073 KFM59278.1 GBXI01013296 JAD00996.1 BDGG01000002 GAU91883.1
KPJ19475.1 MG022107 AYW00836.1 MG022105 AYW00834.1 MG022104 AYW00833.1 MG022106 AYW00835.1 KPJ19472.1 KQ459606 KPI91354.1 MG022101 AYW00830.1 MG022102 AYW00831.1 KPI91353.1 MG022103 AYW00832.1 NWSH01004439 PCG65118.1 ODYU01005911 SOQ47291.1 KPJ19473.1 BABH01027988 MG022099 AYW00828.1 MG022100 AYW00829.1 MG022096 AJ489522 AYW00825.1 CAD33828.1 GFXV01003236 MBW15041.1 AJ410304 CAC86338.1 AJ410305 AJ410306 CAC86342.1 MG022098 AYW00827.1 KX986117 ATE50179.1 KQ971338 EFA02301.1 KK852631 KDR19829.1 NEVH01015305 PNF27140.1 MG022097 AYW00826.1 ABLF02026482 NEVH01004410 PNF39713.1 KX986126 ATE50188.1 GGMS01016567 MBY85770.1 KX986120 ATE50182.1 GEZM01009631 JAV94444.1 KX986122 ATE50184.1 DS235886 EEB20446.1 KZ288227 PBC31752.1 KX986130 ATE50192.1 GFXV01005138 MBW16943.1 GFDF01000380 JAV13704.1 GGMR01011065 MBY23684.1 KQ434910 KZC11351.1 KQ414579 KOC71090.1 GEBQ01002945 JAT37032.1 GGMS01011701 MBY80904.1 KX986121 ATE50183.1 GEDC01030550 JAS06748.1 GFDF01000384 JAV13700.1 KX986124 ATE50186.1 ACPB03014568 ACPB03014569 GFDF01000385 JAV13699.1 KX986115 ATE50177.1 GECZ01012437 JAS57332.1 GEDC01012740 GEDC01012295 JAS24558.1 JAS25003.1 EFA02715.1 KX986118 ATE50180.1 QOIP01000012 RLU15878.1 KQ763209 OAD55144.1 KK852917 KDR13867.1 KX986128 ATE50190.1 UFQS01000723 UFQT01000723 SSX06412.1 SSX26765.1 GBHO01022208 JAG21396.1 GFWV01018204 MAA42932.1 KK852878 KDR14503.1 KK852655 KDR19289.1 GBRD01016667 GDHC01012310 JAG49160.1 JAQ06319.1 GDHC01005360 JAQ13269.1 ODYU01010728 SOQ56015.1 GBRD01016664 GDHC01015135 JAG49163.1 JAQ03494.1 KQ435708 KOX80112.1 MG022110 AYW00839.1 NEVH01025657 PNF15110.1 PNF15112.1 KQ414740 KOC62043.1 ADTU01001092 LJIJ01000470 ODM97123.1 PYGN01000874 PSN39814.1 KDR19827.1 GL435766 EFN72627.1 KQ983214 KYQ46561.1 QOIP01000001 RLU26669.1 DS235882 EEB20157.1 CH964101 EDW79608.1 ACPB03016276 ACPB03016277 KB202481 ESO90113.1 KK113073 KFM59278.1 GBXI01013296 JAD00996.1 BDGG01000002 GAU91883.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000079169
UP000007266
+ More
UP000027135 UP000235965 UP000007819 UP000009046 UP000242457 UP000076502 UP000053825 UP000005203 UP000015103 UP000279307 UP000192223 UP000053105 UP000002358 UP000005205 UP000094527 UP000245037 UP000000311 UP000075809 UP000007798 UP000030746 UP000054359 UP000186922
UP000027135 UP000235965 UP000007819 UP000009046 UP000242457 UP000076502 UP000053825 UP000005203 UP000015103 UP000279307 UP000192223 UP000053105 UP000002358 UP000005205 UP000094527 UP000245037 UP000000311 UP000075809 UP000007798 UP000030746 UP000054359 UP000186922
Interpro
Gene 3D
ProteinModelPortal
H9JSK1
A0A3G5BLW6
A0A3G5BLV4
A0A194RQ78
A0A3G5BLV9
A0A3G5BM44
+ More
A0A3G5BLS9 A0A3G5BLU4 A0A194RPN9 A0A194PF78 A0A3G5BLS3 A0A3G5BLY0 A0A194PE98 A0A3G5BLX5 A0A2A4J0B7 A0A2H1W2J1 A0A194RNQ9 H9JSK0 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 A0A2H8TM77 Q8MPI1 Q8MM72 A0A3G5BLU7 A0A384WE16 A0A3Q0IUC2 D2A3E0 A0A067RK57 A0A2J7QEX8 A0A3G5BLV1 J9KB05 A0A2J7RFU7 A0A384X577 A0A2S2R7V2 A0A384WD78 A0A1Y1N921 A0A384WD96 E0W490 A0A2A3EKI1 A0A384WD97 A0A2H8TRW6 A0A1L8E4Y8 A0A2S2P2T4 A0A154PHM0 A0A0L7RJP2 A0A1B6MM89 A0A2S2QT45 A0A384WDF6 A0A1B6BZR2 A0A1L8E4N6 A0A384WXG8 A0A087ZPU2 T1HN07 A0A1L8E508 A0A384WD64 A0A1B6G4I4 A0A1B6DH44 D2A206 A0A384WT78 A0A3L8D7K7 A0A1W4XG67 A0A310SCZ1 A0A067QUM9 A0A384WT96 A0A087ZZX8 A0A336M966 A0A0A9XP28 A0A293MQ50 A0A067QYG0 A0A067R9K3 A0A0K8S8T4 A0A146M1G8 A0A2H1WSG7 A0A0K8S7A3 A0A0N0BK53 K7J2L9 A0A3G5BLV6 A0A2J7PFJ8 A0A2J7PFK0 A0A0L7QTR2 A0A158P1M3 A0A1W4X523 A0A1D2MVN0 A0A1S3DG86 A0A2P8Y6M3 A0A067R8L8 E2A1F4 A0A151WFD5 A0A3L8E386 E0W3F1 B4N5G9 T1I2L4 V3ZFY3 A0A087T2D9 A0A0A1WQR8 A0A1D1UZJ7
A0A3G5BLS9 A0A3G5BLU4 A0A194RPN9 A0A194PF78 A0A3G5BLS3 A0A3G5BLY0 A0A194PE98 A0A3G5BLX5 A0A2A4J0B7 A0A2H1W2J1 A0A194RNQ9 H9JSK0 A0A3G5BLW2 A0A3G5BLU8 Q8MPH9 A0A2H8TM77 Q8MPI1 Q8MM72 A0A3G5BLU7 A0A384WE16 A0A3Q0IUC2 D2A3E0 A0A067RK57 A0A2J7QEX8 A0A3G5BLV1 J9KB05 A0A2J7RFU7 A0A384X577 A0A2S2R7V2 A0A384WD78 A0A1Y1N921 A0A384WD96 E0W490 A0A2A3EKI1 A0A384WD97 A0A2H8TRW6 A0A1L8E4Y8 A0A2S2P2T4 A0A154PHM0 A0A0L7RJP2 A0A1B6MM89 A0A2S2QT45 A0A384WDF6 A0A1B6BZR2 A0A1L8E4N6 A0A384WXG8 A0A087ZPU2 T1HN07 A0A1L8E508 A0A384WD64 A0A1B6G4I4 A0A1B6DH44 D2A206 A0A384WT78 A0A3L8D7K7 A0A1W4XG67 A0A310SCZ1 A0A067QUM9 A0A384WT96 A0A087ZZX8 A0A336M966 A0A0A9XP28 A0A293MQ50 A0A067QYG0 A0A067R9K3 A0A0K8S8T4 A0A146M1G8 A0A2H1WSG7 A0A0K8S7A3 A0A0N0BK53 K7J2L9 A0A3G5BLV6 A0A2J7PFJ8 A0A2J7PFK0 A0A0L7QTR2 A0A158P1M3 A0A1W4X523 A0A1D2MVN0 A0A1S3DG86 A0A2P8Y6M3 A0A067R8L8 E2A1F4 A0A151WFD5 A0A3L8E386 E0W3F1 B4N5G9 T1I2L4 V3ZFY3 A0A087T2D9 A0A0A1WQR8 A0A1D1UZJ7
PDB
1FSU
E-value=4.6848e-62,
Score=605
Ontologies
GO
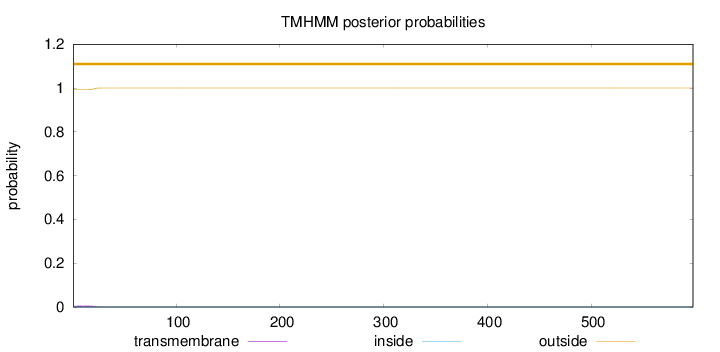
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.970962
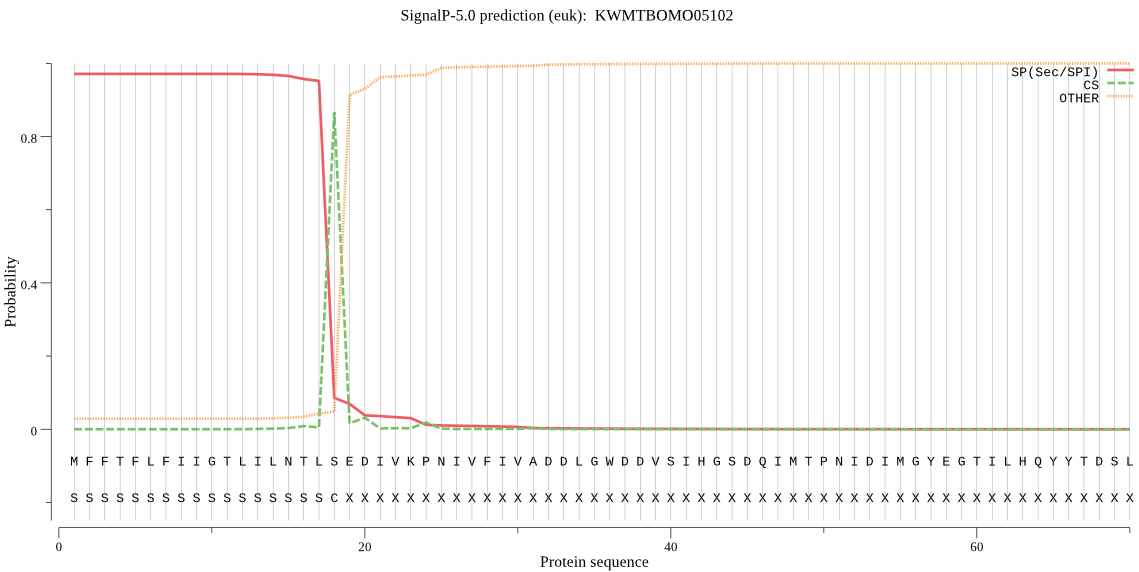
Length:
598
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12019
Exp number, first 60 AAs:
0.11869
Total prob of N-in:
0.00638
outside
1 - 598
Population Genetic Test Statistics
Pi
230.219909
Theta
196.920716
Tajima's D
1.143208
CLR
0.36287
CSRT
0.708564571771411
Interpretation
Uncertain
