Pre Gene Modal
BGIBMGA012514
Annotation
PREDICTED:_tubulin-specific_chaperone_D_[Amyelois_transitella]
Full name
Tubulin-specific chaperone D
Alternative Name
Beta-tubulin cofactor D
SSD-1
Tubulin-folding cofactor D
SSD-1
Tubulin-folding cofactor D
Location in the cell
Cytoplasmic Reliability : 1.895
Sequence
CDS
ATGGTGGGGTTAGACTCTGAGATTAACCGGGATGACGAATGTGATAATATTGGGCTGGGATGTGCACTCGAATACTTTTCAGAGGTCGAGGATGTGATGAACATGATTGATAATGTAAAAAATGTTTATAACACCTCTGCTTTTGAAGTGGAGTACGATAAACTTTACACAATACTTAAACAGTATTATGAACAACCACATCTGTTGGATCCACATTTAGACAAAATTCTATCGAAATTTTTGTTACTTATCAAAGATAAAGATTCTCCTTTCGAACTGAAACATGCAACTTTTAATTACATGTACCAAATCATAAGAGTTCGAGGCTATAAGGTGGTTGTTAGACATCTTCCACATGAAGTGTCTGATTTGCTGACTGTATTGGTATTTTTGGAAGCTCAAAATCCTGATGATAAGGAAACATGGAGAAGCAGATTTGTCTTACTGTTATGGTTGTCAATTGTTGTTATTATTCCATTTCATATGAGTCGCCTAGATGGATTCGCTCCTGGACAACCAGAAGCAGGCACTTCGAAAAAATTAACAGTCATGGAAAGAATCTTTAATATTTGTAAAACATATGCACTTAGCAAAGATTCATGTGCTGAAGCCAGTGCTTATTTAGCCTCTAAATTTTTTACAAGGTCAGATGTGAAAGAATTATATATGGGTGCATTCTTTGATTGGGCCTGTGAGCTGAATTCAAATTTAAATGAAGAGAACACTATACACTACGGGGTTTTAGCTGCTGTTGCAGCTGTTCTGAAGCACGGGAAAAGAGATGATTTACTACCATATGCACCTAAATTACTTGAATGGGTTACGAACCAAAACTATAAGCATCATAATGCTATGCTTGTTAGAAAATATGGAGTTAAAATTGTCCAAAGAATTGGTTTAACATTTCTGCGTCCAAGAGTAGCTGCCTGGCGGTACACTCGTGGTTCAAGGTCTTTAGCGGTTACTTTAGGTGGAGCAGCGGCAGCTGGCGATACGGAAACGATTACTACTCCTGACGATGATGATGATCAAGACATTCCTCAAGAGGTTGAAGATGTAGTAGAACTATTATTGTGTTCACTAAGTGATGACGATACAGTAGTTCGTTGGTCGGCCGCGAAAGGAGTGGGGAGAATCGGTGCACGTTTACCTGCGATAGCCGCTGCTGATGTATGCGAAAGTGTGTTAGGACTGTTTGCAGAAAATGAACGTGAAACTGCCTGGCATGGTGGGTGTATGGCACTGGCTGAACTAGGTAAGTTTTGCTTGTATTGA
Protein
MVGLDSEINRDDECDNIGLGCALEYFSEVEDVMNMIDNVKNVYNTSAFEVEYDKLYTILKQYYEQPHLLDPHLDKILSKFLLLIKDKDSPFELKHATFNYMYQIIRVRGYKVVVRHLPHEVSDLLTVLVFLEAQNPDDKETWRSRFVLLLWLSIVVIIPFHMSRLDGFAPGQPEAGTSKKLTVMERIFNICKTYALSKDSCAEASAYLASKFFTRSDVKELYMGAFFDWACELNSNLNEENTIHYGVLAAVAAVLKHGKRDDLLPYAPKLLEWVTNQNYKHHNAMLVRKYGVKIVQRIGLTFLRPRVAAWRYTRGSRSLAVTLGGAAAAGDTETITTPDDDDDQDIPQEVEDVVELLLCSLSDDDTVVRWSAAKGVGRIGARLPAIAAADVCESVLGLFAENERETAWHGGCMALAELGKFCLY
Summary
Description
Tubulin-folding protein implicated in the first step of the tubulin folding pathway and required for tubulin complex assembly. Involved in the regulation of microtubule polymerization or depolymerization, it modulates microtubule dynamics by capturing GTP-bound beta-tubulin (TUBB). Its ability to interact with beta tubulin is regulated via its interaction with ARL2. Acts as a GTPase-activating protein (GAP) for ARL2. Induces microtubule disruption in absence of ARL2. Increases degradation of beta tubulin, when overexpressed in polarized cells. Promotes epithelial cell detachment, a process antagonized by ARL2. Induces tight adherens and tight junctions disassembly at the lateral cell membrane (PubMed:10722852, PubMed:10831612, PubMed:11847227, PubMed:20740604, PubMed:27666370, PubMed:28158450). Required for correct assembly and maintenance of the mitotic spindle, and proper progression of mitosis (PubMed:27666370). Involved in neuron morphogenesis (PubMed:27666374).
Tubulin-folding protein implicated in the first step of the tubulin folding pathway and required for tubulin complex assembly. Involved in the regulation of microtubule polymerization or depolymerization, it modulates microtubule dynamics by capturing GTP-bound beta-tubulin (TUBB). Its ability to interact with beta tubulin is regulated via its interaction with ARL2. Acts as a GTPase-activating protein (GAP) for ARL2. Induces microtubule disruption in absence of ARL2. Increases degradation of beta tubulin, when overexpressed in polarized cells. Promotes epithelial cell detachment, a process antagonized by ARL2. Induces tight adherens and tight junctions disassembly at the lateral cell membrane. Required for correct assembly and maintenance of the mitotic spindle, and proper progression of mitosis. Involved in neuron morphogenesis.
Tubulin-folding protein implicated in the first step of the tubulin folding pathway and required for tubulin complex assembly. Involved in the regulation of microtubule polymerization or depolymerization, it modulates microtubule dynamics by capturing GTP-bound beta-tubulin (TUBB). Its ability to interact with beta tubulin is regulated via its interaction with ARL2. Acts as a GTPase-activating protein (GAP) for ARL2. Induces microtubule disruption in absence of ARL2. Increases degradation of beta tubulin, when overexpressed in polarized cells. Promotes epithelial cell detachment, a process antagonized by ARL2. Induces tight adherens and tight junctions disassembly at the lateral cell membrane. Required for correct assembly and maintenance of the mitotic spindle, and proper progression of mitosis. Involved in neuron morphogenesis.
Subunit
Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with PPP2CB (By similarity). Part of a supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state (PubMed:10831612). Interacts with ARL2; interaction is enhanced with the GDP-bound form of ARL2 (PubMed:10831612, PubMed:27666374). Does not interact with ARL3, ARL4A and ARL4D (PubMed:10831612). Interacts with beta tubulin (PubMed:10831612, PubMed:27666370, PubMed:27666374). Interacts with TBCE (PubMed:27666374).
Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with PPP2CB (By similarity). Part of a supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Interacts with ARL2; interaction is enhanced with the GDP-bound form of ARL2. Does not interact with ARL3, ARL4A and ARL4D. Interacts with beta tubulin. Interacts with TBCE (By similarity).
Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with PPP2CB. Part of a supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Interacts with ARL2; interaction is enhanced with the GDP-bound form of ARL2. Does not interact with ARL3, ARL4A and ARL4D. Interacts with beta tubulin. Interacts with TBCE (By similarity) (PubMed:12912990, PubMed:17704193).
Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with PPP2CB (By similarity). Part of a supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Interacts with ARL2; interaction is enhanced with the GDP-bound form of ARL2. Does not interact with ARL3, ARL4A and ARL4D. Interacts with beta tubulin. Interacts with TBCE (By similarity).
Found in a complex with at least ARL2, PPP2CB, PPP2R1A, PPP2R2A, PPP2R5E and TBCD. Interacts with PPP2CB. Part of a supercomplex made of cofactors A to E. Cofactors A and D function by capturing and stabilizing tubulin in a quasi-native conformation. Cofactor E binds to the cofactor D-tubulin complex; interaction with cofactor C then causes the release of tubulin polypeptides that are committed to the native state. Interacts with ARL2; interaction is enhanced with the GDP-bound form of ARL2. Does not interact with ARL3, ARL4A and ARL4D. Interacts with beta tubulin. Interacts with TBCE (By similarity) (PubMed:12912990, PubMed:17704193).
Similarity
Belongs to the TBCD family.
Keywords
Alternative splicing
Cell junction
Cell membrane
Chaperone
Complete proteome
Cytoplasm
Cytoskeleton
Disease mutation
GTPase activation
Membrane
Neurodegeneration
Polymorphism
Reference proteome
Repeat
Tight junction
Feature
chain Tubulin-specific chaperone D
splice variant In isoform 2.
sequence variant In PEBAT; dbSNP:rs778417127.
splice variant In isoform 2.
sequence variant In PEBAT; dbSNP:rs778417127.
Uniprot
A0A2H1WTB1
A0A2A4J981
A0A212EIN0
A0A194PE89
A0A194RPP8
A0A1E1WTE3
+ More
U4UGW6 A0A2J7RTG4 A0A1Y1MM56 A0A1W4WQ67 D6X4J7 A0A2J7RTH2 A0A1B6E9Z8 E0VS67 B4Q7M9 B4IM58 B4NW50 B3MJC5 Q9VQ78 A0A1W4V7E9 B3N9G0 A0A2P6L8N9 A0A1J1IW54 A0A2Y9DGV0 I6L959 A0A1A9WDM9 H0XAQ4 A0A2U9CR13 A0A087UCN3 B4DE53 A0A2K6EYK2 A0A2U9CR32 A0A1B0FQX1 F7ACU9 G1RIG4 A0A2K6EYL0 B4JAR1 A0A2I3HHJ3 A0A2U9CTN0 A0A1S3WCX2 A0A2U9CT71 A0A1B0A5Y4 A0A1A9X610 F7B7Q7 A0A1B0BWC5 H2NV72 A0A0M4ECN4 K7GD88 K7GD94 A0A0A1X877 A0A210QA62 A0A3Q2CP16 K1RAT9 A0A3Q7UH91 A0A3Q2E073 Q9BTW9 A0A3Q2FPB8 A0A2Y9GGB9 J3KR97 A0A1A9V6V7 A0A3Q7V9E0 Q9BTW9-4 F1NK98 A0A3Q7PHT3 A0A151NVM2 Q5ZI87 A0A340XCW2 K7BVH1 H0VEQ7 E1BU18 H2QE60 A0A2J8JF69 A0A2I3TB15 A0A1D5NTW7 A0A2Y9LCT3 A0A3P4MX50 A0A3B4A0V0 A0A2Y9L6T5 W5Q3B4 A0A3B0JAG8 A0A1A8FFT3 A0A3B4UPJ5 A0A2Y9FKF1 A0A2K5P636 A0A3Q2PHB4 Q0V9L2 A7MB50 A0A0S3NT15 H9YV54 H9FZ08 I0FKI9 A0A3S2M7Q0 A0A096NTQ7 A0A2K5P698 A0A2U3W8J7 L8J3R2 Q28205 A0A384AF66 A0A1A8BX37
U4UGW6 A0A2J7RTG4 A0A1Y1MM56 A0A1W4WQ67 D6X4J7 A0A2J7RTH2 A0A1B6E9Z8 E0VS67 B4Q7M9 B4IM58 B4NW50 B3MJC5 Q9VQ78 A0A1W4V7E9 B3N9G0 A0A2P6L8N9 A0A1J1IW54 A0A2Y9DGV0 I6L959 A0A1A9WDM9 H0XAQ4 A0A2U9CR13 A0A087UCN3 B4DE53 A0A2K6EYK2 A0A2U9CR32 A0A1B0FQX1 F7ACU9 G1RIG4 A0A2K6EYL0 B4JAR1 A0A2I3HHJ3 A0A2U9CTN0 A0A1S3WCX2 A0A2U9CT71 A0A1B0A5Y4 A0A1A9X610 F7B7Q7 A0A1B0BWC5 H2NV72 A0A0M4ECN4 K7GD88 K7GD94 A0A0A1X877 A0A210QA62 A0A3Q2CP16 K1RAT9 A0A3Q7UH91 A0A3Q2E073 Q9BTW9 A0A3Q2FPB8 A0A2Y9GGB9 J3KR97 A0A1A9V6V7 A0A3Q7V9E0 Q9BTW9-4 F1NK98 A0A3Q7PHT3 A0A151NVM2 Q5ZI87 A0A340XCW2 K7BVH1 H0VEQ7 E1BU18 H2QE60 A0A2J8JF69 A0A2I3TB15 A0A1D5NTW7 A0A2Y9LCT3 A0A3P4MX50 A0A3B4A0V0 A0A2Y9L6T5 W5Q3B4 A0A3B0JAG8 A0A1A8FFT3 A0A3B4UPJ5 A0A2Y9FKF1 A0A2K5P636 A0A3Q2PHB4 Q0V9L2 A7MB50 A0A0S3NT15 H9YV54 H9FZ08 I0FKI9 A0A3S2M7Q0 A0A096NTQ7 A0A2K5P698 A0A2U3W8J7 L8J3R2 Q28205 A0A384AF66 A0A1A8BX37
Pubmed
22118469
26354079
23537049
28004739
18362917
19820115
+ More
20566863 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15489334 17495919 18464734 17381049 25830018 28812685 22992520 11110777 15498874 10231032 14702039 17974005 16625196 10722852 10831612 11847227 20740604 21269460 27666374 27666370 28158450 27807845 15592404 22293439 15642098 21993624 16136131 25463417 20809919 26514418 25319552 22751099 8706133 12912990 17704193
20566863 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15489334 17495919 18464734 17381049 25830018 28812685 22992520 11110777 15498874 10231032 14702039 17974005 16625196 10722852 10831612 11847227 20740604 21269460 27666374 27666370 28158450 27807845 15592404 22293439 15642098 21993624 16136131 25463417 20809919 26514418 25319552 22751099 8706133 12912990 17704193
EMBL
ODYU01010899
SOQ56293.1
NWSH01002295
PCG68635.1
AGBW02014611
OWR41346.1
+ More
KQ459606 KPI91343.1 KQ459896 KPJ19482.1 GDQN01000948 JAT90106.1 KB632350 ERL93229.1 NEVH01000003 PNF44135.1 GEZM01031220 JAV84966.1 KQ971380 EEZ97256.1 PNF44134.1 GEDC01002555 JAS34743.1 DS235745 EEB16223.1 CM000361 CM002910 EDX03409.1 KMY87586.1 CH480936 EDW44538.1 CM000157 EDW87330.1 CH902620 EDV31335.1 AE014134 BT031324 AAF51300.1 ABY21737.1 CH954177 EDV57417.1 MWRG01000953 PRD34941.1 CVRI01000063 CRL04336.1 BC006364 AAH06364.1 AAQR03040706 AAQR03040707 AAQR03040708 AAQR03040709 AAQR03040710 AAQR03040711 AAQR03040712 AAQR03040713 AAQR03040714 AAQR03040715 CP026261 AWP19028.1 KK119224 KFM75122.1 AK293471 BAG56964.1 AWP19027.1 CCAG010010711 ADFV01131713 ADFV01131714 ADFV01131715 ADFV01131716 ADFV01131717 ADFV01131718 ADFV01131719 ADFV01131720 ADFV01131721 ADFV01131722 CH916368 EDW03869.1 AWP19026.1 AWP19029.1 JXJN01021708 ABGA01291964 ABGA01326382 ABGA01326383 ABGA01326384 ABGA01326385 ABGA01326386 ABGA01326387 ABGA01326388 ABGA01326389 ABGA01326390 CP012523 ALC39422.1 AGCU01056393 AGCU01056394 AGCU01056395 AGCU01056396 AGCU01056397 AGCU01056398 AGCU01056399 AGCU01056400 AGCU01056401 AGCU01056402 GBXI01007005 JAD07287.1 NEDP02004422 OWF45618.1 JH823244 EKC31146.1 AJ006417 AF193042 AB023205 AK091959 AL133562 AL096745 AC024361 AC068014 AC068584 AC087222 AC130371 BC003094 BC012824 BC039654 AADN05000628 AKHW03001857 KYO40838.1 AJ720897 GABC01009260 GABF01006145 GABD01006625 GABE01008072 NBAG03000462 JAA02078.1 JAA16000.1 JAA26475.1 JAA36667.1 PNI21406.1 AAKN02047363 AAKN02047364 AACZ04058693 AACZ04058694 AACZ04058695 AACZ04058696 AACZ04058697 AACZ04058698 AACZ04058699 AC192631 PNI21407.1 CYRY02015426 VCW85437.1 AMGL01018993 AMGL01018994 AMGL01018995 AMGL01018996 OUUW01000004 SPP79297.1 HAEB01011355 SBQ57882.1 BC121491 AAI21492.1 BC151338 AAI51339.1 AB984629 BAT46490.1 JU470766 AFH27570.1 JU336114 AFE79867.1 JV044894 AFI34965.1 CM012444 RVE69637.1 AHZZ02010864 AHZZ02010865 AHZZ02010866 JH880371 ELR62234.1 U61233 HADZ01007119 HAEA01009026 SBP71060.1
KQ459606 KPI91343.1 KQ459896 KPJ19482.1 GDQN01000948 JAT90106.1 KB632350 ERL93229.1 NEVH01000003 PNF44135.1 GEZM01031220 JAV84966.1 KQ971380 EEZ97256.1 PNF44134.1 GEDC01002555 JAS34743.1 DS235745 EEB16223.1 CM000361 CM002910 EDX03409.1 KMY87586.1 CH480936 EDW44538.1 CM000157 EDW87330.1 CH902620 EDV31335.1 AE014134 BT031324 AAF51300.1 ABY21737.1 CH954177 EDV57417.1 MWRG01000953 PRD34941.1 CVRI01000063 CRL04336.1 BC006364 AAH06364.1 AAQR03040706 AAQR03040707 AAQR03040708 AAQR03040709 AAQR03040710 AAQR03040711 AAQR03040712 AAQR03040713 AAQR03040714 AAQR03040715 CP026261 AWP19028.1 KK119224 KFM75122.1 AK293471 BAG56964.1 AWP19027.1 CCAG010010711 ADFV01131713 ADFV01131714 ADFV01131715 ADFV01131716 ADFV01131717 ADFV01131718 ADFV01131719 ADFV01131720 ADFV01131721 ADFV01131722 CH916368 EDW03869.1 AWP19026.1 AWP19029.1 JXJN01021708 ABGA01291964 ABGA01326382 ABGA01326383 ABGA01326384 ABGA01326385 ABGA01326386 ABGA01326387 ABGA01326388 ABGA01326389 ABGA01326390 CP012523 ALC39422.1 AGCU01056393 AGCU01056394 AGCU01056395 AGCU01056396 AGCU01056397 AGCU01056398 AGCU01056399 AGCU01056400 AGCU01056401 AGCU01056402 GBXI01007005 JAD07287.1 NEDP02004422 OWF45618.1 JH823244 EKC31146.1 AJ006417 AF193042 AB023205 AK091959 AL133562 AL096745 AC024361 AC068014 AC068584 AC087222 AC130371 BC003094 BC012824 BC039654 AADN05000628 AKHW03001857 KYO40838.1 AJ720897 GABC01009260 GABF01006145 GABD01006625 GABE01008072 NBAG03000462 JAA02078.1 JAA16000.1 JAA26475.1 JAA36667.1 PNI21406.1 AAKN02047363 AAKN02047364 AACZ04058693 AACZ04058694 AACZ04058695 AACZ04058696 AACZ04058697 AACZ04058698 AACZ04058699 AC192631 PNI21407.1 CYRY02015426 VCW85437.1 AMGL01018993 AMGL01018994 AMGL01018995 AMGL01018996 OUUW01000004 SPP79297.1 HAEB01011355 SBQ57882.1 BC121491 AAI21492.1 BC151338 AAI51339.1 AB984629 BAT46490.1 JU470766 AFH27570.1 JU336114 AFE79867.1 JV044894 AFI34965.1 CM012444 RVE69637.1 AHZZ02010864 AHZZ02010865 AHZZ02010866 JH880371 ELR62234.1 U61233 HADZ01007119 HAEA01009026 SBP71060.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000030742
UP000235965
+ More
UP000192223 UP000007266 UP000009046 UP000000304 UP000001292 UP000002282 UP000007801 UP000000803 UP000192221 UP000008711 UP000183832 UP000248480 UP000091820 UP000005225 UP000246464 UP000054359 UP000233160 UP000092444 UP000002280 UP000001073 UP000001070 UP000079721 UP000092445 UP000092443 UP000002279 UP000092460 UP000001595 UP000092553 UP000007267 UP000242188 UP000265020 UP000005408 UP000286642 UP000005640 UP000248481 UP000078200 UP000000539 UP000286641 UP000050525 UP000265300 UP000005447 UP000002277 UP000248482 UP000261520 UP000002356 UP000268350 UP000261420 UP000233060 UP000265000 UP000028761 UP000245340 UP000009136 UP000261681
UP000192223 UP000007266 UP000009046 UP000000304 UP000001292 UP000002282 UP000007801 UP000000803 UP000192221 UP000008711 UP000183832 UP000248480 UP000091820 UP000005225 UP000246464 UP000054359 UP000233160 UP000092444 UP000002280 UP000001073 UP000001070 UP000079721 UP000092445 UP000092443 UP000002279 UP000092460 UP000001595 UP000092553 UP000007267 UP000242188 UP000265020 UP000005408 UP000286642 UP000005640 UP000248481 UP000078200 UP000000539 UP000286641 UP000050525 UP000265300 UP000005447 UP000002277 UP000248482 UP000261520 UP000002356 UP000268350 UP000261420 UP000233060 UP000265000 UP000028761 UP000245340 UP000009136 UP000261681
Pfam
PF12612 TFCD_C
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2H1WTB1
A0A2A4J981
A0A212EIN0
A0A194PE89
A0A194RPP8
A0A1E1WTE3
+ More
U4UGW6 A0A2J7RTG4 A0A1Y1MM56 A0A1W4WQ67 D6X4J7 A0A2J7RTH2 A0A1B6E9Z8 E0VS67 B4Q7M9 B4IM58 B4NW50 B3MJC5 Q9VQ78 A0A1W4V7E9 B3N9G0 A0A2P6L8N9 A0A1J1IW54 A0A2Y9DGV0 I6L959 A0A1A9WDM9 H0XAQ4 A0A2U9CR13 A0A087UCN3 B4DE53 A0A2K6EYK2 A0A2U9CR32 A0A1B0FQX1 F7ACU9 G1RIG4 A0A2K6EYL0 B4JAR1 A0A2I3HHJ3 A0A2U9CTN0 A0A1S3WCX2 A0A2U9CT71 A0A1B0A5Y4 A0A1A9X610 F7B7Q7 A0A1B0BWC5 H2NV72 A0A0M4ECN4 K7GD88 K7GD94 A0A0A1X877 A0A210QA62 A0A3Q2CP16 K1RAT9 A0A3Q7UH91 A0A3Q2E073 Q9BTW9 A0A3Q2FPB8 A0A2Y9GGB9 J3KR97 A0A1A9V6V7 A0A3Q7V9E0 Q9BTW9-4 F1NK98 A0A3Q7PHT3 A0A151NVM2 Q5ZI87 A0A340XCW2 K7BVH1 H0VEQ7 E1BU18 H2QE60 A0A2J8JF69 A0A2I3TB15 A0A1D5NTW7 A0A2Y9LCT3 A0A3P4MX50 A0A3B4A0V0 A0A2Y9L6T5 W5Q3B4 A0A3B0JAG8 A0A1A8FFT3 A0A3B4UPJ5 A0A2Y9FKF1 A0A2K5P636 A0A3Q2PHB4 Q0V9L2 A7MB50 A0A0S3NT15 H9YV54 H9FZ08 I0FKI9 A0A3S2M7Q0 A0A096NTQ7 A0A2K5P698 A0A2U3W8J7 L8J3R2 Q28205 A0A384AF66 A0A1A8BX37
U4UGW6 A0A2J7RTG4 A0A1Y1MM56 A0A1W4WQ67 D6X4J7 A0A2J7RTH2 A0A1B6E9Z8 E0VS67 B4Q7M9 B4IM58 B4NW50 B3MJC5 Q9VQ78 A0A1W4V7E9 B3N9G0 A0A2P6L8N9 A0A1J1IW54 A0A2Y9DGV0 I6L959 A0A1A9WDM9 H0XAQ4 A0A2U9CR13 A0A087UCN3 B4DE53 A0A2K6EYK2 A0A2U9CR32 A0A1B0FQX1 F7ACU9 G1RIG4 A0A2K6EYL0 B4JAR1 A0A2I3HHJ3 A0A2U9CTN0 A0A1S3WCX2 A0A2U9CT71 A0A1B0A5Y4 A0A1A9X610 F7B7Q7 A0A1B0BWC5 H2NV72 A0A0M4ECN4 K7GD88 K7GD94 A0A0A1X877 A0A210QA62 A0A3Q2CP16 K1RAT9 A0A3Q7UH91 A0A3Q2E073 Q9BTW9 A0A3Q2FPB8 A0A2Y9GGB9 J3KR97 A0A1A9V6V7 A0A3Q7V9E0 Q9BTW9-4 F1NK98 A0A3Q7PHT3 A0A151NVM2 Q5ZI87 A0A340XCW2 K7BVH1 H0VEQ7 E1BU18 H2QE60 A0A2J8JF69 A0A2I3TB15 A0A1D5NTW7 A0A2Y9LCT3 A0A3P4MX50 A0A3B4A0V0 A0A2Y9L6T5 W5Q3B4 A0A3B0JAG8 A0A1A8FFT3 A0A3B4UPJ5 A0A2Y9FKF1 A0A2K5P636 A0A3Q2PHB4 Q0V9L2 A7MB50 A0A0S3NT15 H9YV54 H9FZ08 I0FKI9 A0A3S2M7Q0 A0A096NTQ7 A0A2K5P698 A0A2U3W8J7 L8J3R2 Q28205 A0A384AF66 A0A1A8BX37
Ontologies
GO
GO:0048487
GO:0016021
GO:0005096
GO:0007023
GO:0007021
GO:0006457
GO:0070830
GO:0005923
GO:0000226
GO:0005912
GO:0034333
GO:0016328
GO:0007409
GO:1902850
GO:0045196
GO:0048813
GO:0031115
GO:0043547
GO:0005813
GO:0000278
GO:0048667
GO:0007420
GO:0010842
GO:0005874
GO:0010812
GO:0005737
GO:0051087
GO:0005815
GO:0005829
GO:0005488
GO:0007186
GO:0016020
GO:0004888
GO:0007166
GO:0016616
GO:0046168
GO:0016491
GO:0003824
PANTHER
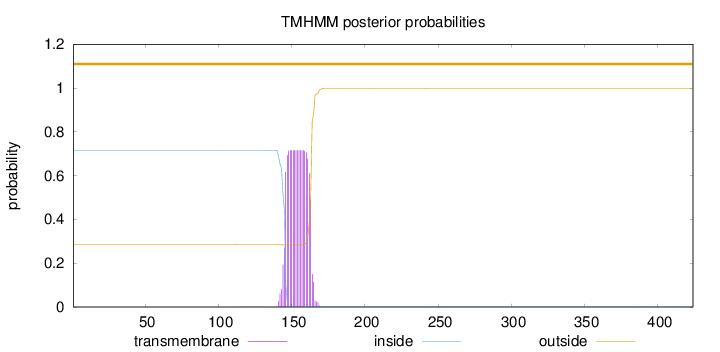
Topology
Subcellular location
Cell junction
Localized in cell-cell contacts. With evidence from 1 publications.
Tight junction Localized in cell-cell contacts. With evidence from 1 publications.
Lateral cell membrane Localized in cell-cell contacts. With evidence from 1 publications.
Cytoplasm Localized in cell-cell contacts. With evidence from 1 publications.
Adherens junction Localized in cell-cell contacts. With evidence from 1 publications.
Cytoskeleton Localized in cell-cell contacts. With evidence from 1 publications.
Microtubule organizing center Localized in cell-cell contacts. With evidence from 1 publications.
Centrosome Localized in cell-cell contacts. With evidence from 1 publications.
Tight junction Localized in cell-cell contacts. With evidence from 1 publications.
Lateral cell membrane Localized in cell-cell contacts. With evidence from 1 publications.
Cytoplasm Localized in cell-cell contacts. With evidence from 1 publications.
Adherens junction Localized in cell-cell contacts. With evidence from 1 publications.
Cytoskeleton Localized in cell-cell contacts. With evidence from 1 publications.
Microtubule organizing center Localized in cell-cell contacts. With evidence from 1 publications.
Centrosome Localized in cell-cell contacts. With evidence from 1 publications.
Length:
424
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.38979
Exp number, first 60 AAs:
0
Total prob of N-in:
0.71502
outside
1 - 424
Population Genetic Test Statistics
Pi
270.813441
Theta
176.752927
Tajima's D
2.066023
CLR
0.248605
CSRT
0.891305434728264
Interpretation
Uncertain
