Gene
KWMTBOMO05097 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012514
Annotation
PREDICTED:_tubulin-specific_chaperone_D_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.965
Sequence
CDS
ATGCTCGCCAACGCACTCATAGCGACAGCTTGTTTTGATCGAGAGATCAATTGCCGTCGAGCTGCATCGGCGGCGTATCAAGAAAATGTGGGACGTCATGGTATGTTCCCGCACGGCATCAATGTGCTCACAACTGCTGACTTTCAATCTGTGGGACCTAGGAACAATGCTTATCTTGTAATAGCCCCACAGATCGCTGAATATCCGGAGTACACACAGCCATTGGTCGATCATCTGGTGGAGCTGAAAGTCGATCATTGGGACTGTGCCATACGCGAACTCGCCGCCAAGGCATTGAACAAACTAACGATTAAGCTTCCGGAATACATGGCAGTGACAGTTTTACCAAAACTATTAAAAAAGACGGAGTCTATCGATTTGAACGTACGACACGGAGCAATACTGGCCATAGGTGAAACTATATACGCATTGTCACAAACAAAATTGCCCAATGGTAAAACAGCAGACAGCTTAATATCCGAGGACATCTCGACATCAGTACGTGAGCTGGTGGCGCGGTTGAGGTCCCGCCAGCAGTTCCGCGGTCTCGGAGGGGAACTGATGCGGCAGGCTTGCTGCAACTGTATAGCGTCGCTTTCGTTGGCCGCAGCACCTTACCACGGACACCCTATTATAGATGAATGGCTAAATTTAATTGAAGAATGTCTAGCTCACGAAGTGCTAGCCATAAGAGAGAAAGCAATAAAAGCTCTGCCATTGGTTTTCGATCAGTATCTTCGAGACGACCAATTGGTGTATGGTGATATGACAGCGAAAGTAAAACGCACTAAACTTATGGAGAAGTACTGCGAGCAACTTAGGAACAGTGGAGTCAATGGACTTGTGCTGAGAATGGGATACTCGCGAGCTGTTGGCGCTTTACCTAAGTTTTTGCTGACCGAACATCTCCCGATGGTCATTCGTTCGCTGATCGAATGCACGAAAGTGACGGAAGCGACGCAGAAGTGGGCGGAGGCGCGTCGCGACGCCGTCATAGGGCTAACTGAAGTGTGCCAGACGCAGGGGGTGCGGGGGGGCATCGAGGAACACGTGGACGATGTCGTGGCTGCACTCTTGGAGTGCTTAGCTGAATATACTATAGATATGAGGGGCGACATAGGCGCTTGGGTCCGGGAAGCCAGTATGTCAGGTCTACTCGCGATATGTCGACAGTGTGCCGAAGAAGCCCCGCAGTTGAACTCTCCACAAATTATCAGAGACGTGATGTGTGGCGTCGCGCAGCAGGCCGTCGAGAAAATTGACCGAACCAGAGCCCACGCGGGACGGATATTCACATCTCTCATATACAACGATCCTCCGATCTCGAACATTCCGCATCACGAAGCATTGAAACGCATATTTCCGTCCGAAGAAGTCGAGCTGAAGCCGCATCCTAAAGATATTGACGATGAAAAAGATCTGTTAACAAGCGAAAACTCAAACGTGGTGCTCTGGTTGTTCCCCGGACACACAATGCCTAGATTTGTTCAGTTATTGAACTATCCAGATTACAGGTACAGTGTGATAAAAGGACTGGTTGTCAGTGCCGGAGAGCTGACCGAGAGCTTGGTAAAACACACAACGCAATCTCTATATACGTATCTGAACACGCTGCATTCGGACAAGGAAATGCTAAAGTCTATTTGCGAGACGATCATCAAAGTTTTTGCCGATAATTTGCATGTGAAACGCATAACGGGACCGATGTTCAACTTTTTGGATCGATTATTGAGTTCAGGTTCCATATCTCCTATCCTGGAAGATCCACAGTCGAGTTTCGCGATGGACGTTCTCAAACATCTGCAGTTGGAATTACGAGGGGGCAAAAATATATACAAGCTGCTAGATTCCATAAATGTCCTTTGTCAATTAATTCAGGTCGGAGGCGGCGTCTGCAGCAAGTCCCTGGGTCAGCTCGTCATCTACCTGTGCTACGCGGACCGCTACGTGCGGCGGTGCGCGGCCGCCAGGCTCTACGAGGCGCTCACGCTCTACGGAGACGTGTGCTCCGTGCCGCAAGACAACATAGACCAGGTGATGTCCGTACTCGCTGAAACCGATTGGGAGCAAGACGTTTCCAAACTGAGGCCTATAAGAAACGAACTCTGCGATCTGATGGACATCAAACGTCCCGTGATGAAGCAAAAATAA
Protein
MLANALIATACFDREINCRRAASAAYQENVGRHGMFPHGINVLTTADFQSVGPRNNAYLVIAPQIAEYPEYTQPLVDHLVELKVDHWDCAIRELAAKALNKLTIKLPEYMAVTVLPKLLKKTESIDLNVRHGAILAIGETIYALSQTKLPNGKTADSLISEDISTSVRELVARLRSRQQFRGLGGELMRQACCNCIASLSLAAAPYHGHPIIDEWLNLIEECLAHEVLAIREKAIKALPLVFDQYLRDDQLVYGDMTAKVKRTKLMEKYCEQLRNSGVNGLVLRMGYSRAVGALPKFLLTEHLPMVIRSLIECTKVTEATQKWAEARRDAVIGLTEVCQTQGVRGGIEEHVDDVVAALLECLAEYTIDMRGDIGAWVREASMSGLLAICRQCAEEAPQLNSPQIIRDVMCGVAQQAVEKIDRTRAHAGRIFTSLIYNDPPISNIPHHEALKRIFPSEEVELKPHPKDIDDEKDLLTSENSNVVLWLFPGHTMPRFVQLLNYPDYRYSVIKGLVVSAGELTESLVKHTTQSLYTYLNTLHSDKEMLKSICETIIKVFADNLHVKRITGPMFNFLDRLLSSGSISPILEDPQSSFAMDVLKHLQLELRGGKNIYKLLDSINVLCQLIQVGGGVCSKSLGQLVIYLCYADRYVRRCAAARLYEALTLYGDVCSVPQDNIDQVMSVLAETDWEQDVSKLRPIRNELCDLMDIKRPVMKQK
Summary
Uniprot
H9JSK3
A0A2H1WTB1
A0A2A4J981
A0A194PE89
A0A212EIN0
A0A194RNR9
+ More
A0A2P8ZKA7 A0A2J7RTG4 A0A2J7RTH2 A0A1Y1MM56 A0A1W4WQ67 A0A1B6E9Z8 D6X4J7 E2AAI9 U4UGW6 A0A0L7R7M2 A0A154PBN3 A0A088AJI0 A0A224XE77 A0A026WLQ2 A0A3L8DXL5 A0A069DY10 A0A195FST5 A0A336LLZ4 E0VS67 A0A158NSH8 F4WFL2 T1HW65 A0A1J1IW54 A0A2A3E6R3 A0A0M8ZQ24 A0A151HZF5 A0A195CTU5 A0A151J4C1 A0A151WI90 E2BW23 A0A0C9PME1 W8BZN8 W8CBY4 A0A034V7C1 A0A034V548 A0A1I8PLG6 A0A1B0D4Y0 A0A0A1X877 A0A1V4KWK5 A0A0K8VYF4 A0A2I0M7H9 A0A210QA62 A0A093PSQ2 A0A0Q3URE3 A0A091LTC1 U3JDB9 A0A091PX27 A0A0L0CG22 A0A218UJW0 A0A1I8M207 T1PKW7 A0A091UP09 A0A091S7W3 H0ZB90 A0A1A9X610 A0A1S3K523 A0A093G5C7 A0A1A9WDM9 A0A151NVM2 A0A1U7RZZ8 A0A1A9V6V7 A0A091KGS2 A0A091H2M9 A0A1B0BWC5 A0A091LEG0 A0A093EU46 V9K9Q8 A0A1B0A5Y4 A0A1D5NTW7 E1BU18 U3ITR5 G1KU24 A0A1B0FQX1 K7GD88 K7IW10 A0A091F635 B3MJC5 A0A232FCX9 A0A093FTT0 F7GUM4 G1MX06 B4LTU3 A0A091WIR9 A0A3Q0CJ53 A0A2B4SLY4 A0A1S3WCX2 G7NHJ1 F1M1D5 A0A0D9S509 G3I449 W5N9F6 H9FZ08 A0A093J268 H9YV54
A0A2P8ZKA7 A0A2J7RTG4 A0A2J7RTH2 A0A1Y1MM56 A0A1W4WQ67 A0A1B6E9Z8 D6X4J7 E2AAI9 U4UGW6 A0A0L7R7M2 A0A154PBN3 A0A088AJI0 A0A224XE77 A0A026WLQ2 A0A3L8DXL5 A0A069DY10 A0A195FST5 A0A336LLZ4 E0VS67 A0A158NSH8 F4WFL2 T1HW65 A0A1J1IW54 A0A2A3E6R3 A0A0M8ZQ24 A0A151HZF5 A0A195CTU5 A0A151J4C1 A0A151WI90 E2BW23 A0A0C9PME1 W8BZN8 W8CBY4 A0A034V7C1 A0A034V548 A0A1I8PLG6 A0A1B0D4Y0 A0A0A1X877 A0A1V4KWK5 A0A0K8VYF4 A0A2I0M7H9 A0A210QA62 A0A093PSQ2 A0A0Q3URE3 A0A091LTC1 U3JDB9 A0A091PX27 A0A0L0CG22 A0A218UJW0 A0A1I8M207 T1PKW7 A0A091UP09 A0A091S7W3 H0ZB90 A0A1A9X610 A0A1S3K523 A0A093G5C7 A0A1A9WDM9 A0A151NVM2 A0A1U7RZZ8 A0A1A9V6V7 A0A091KGS2 A0A091H2M9 A0A1B0BWC5 A0A091LEG0 A0A093EU46 V9K9Q8 A0A1B0A5Y4 A0A1D5NTW7 E1BU18 U3ITR5 G1KU24 A0A1B0FQX1 K7GD88 K7IW10 A0A091F635 B3MJC5 A0A232FCX9 A0A093FTT0 F7GUM4 G1MX06 B4LTU3 A0A091WIR9 A0A3Q0CJ53 A0A2B4SLY4 A0A1S3WCX2 G7NHJ1 F1M1D5 A0A0D9S509 G3I449 W5N9F6 H9FZ08 A0A093J268 H9YV54
Pubmed
19121390
26354079
22118469
29403074
28004739
18362917
+ More
19820115 20798317 23537049 24508170 30249741 26334808 20566863 21347285 21719571 24495485 25348373 25830018 23371554 28812685 26108605 25315136 20360741 22293439 24402279 15592404 23749191 17381049 20075255 17994087 28648823 17431167 20838655 22002653 15057822 21804562 25319552
19820115 20798317 23537049 24508170 30249741 26334808 20566863 21347285 21719571 24495485 25348373 25830018 23371554 28812685 26108605 25315136 20360741 22293439 24402279 15592404 23749191 17381049 20075255 17994087 28648823 17431167 20838655 22002653 15057822 21804562 25319552
EMBL
BABH01028019
BABH01028020
BABH01028021
BABH01028022
BABH01028023
BABH01028024
+ More
BABH01028025 BABH01028026 ODYU01010899 SOQ56293.1 NWSH01002295 PCG68635.1 KQ459606 KPI91343.1 AGBW02014611 OWR41346.1 KQ459896 KPJ19483.1 PYGN01000031 PSN56928.1 NEVH01000003 PNF44135.1 PNF44134.1 GEZM01031220 JAV84966.1 GEDC01002555 JAS34743.1 KQ971380 EEZ97256.1 GL438128 EFN69536.1 KB632350 ERL93229.1 KQ414642 KOC66771.1 KQ434869 KZC09295.1 GFTR01008328 JAW08098.1 KK107161 EZA56576.1 QOIP01000003 RLU25214.1 GBGD01000173 JAC88716.1 KQ981276 KYN43508.1 UFQT01000015 SSX17771.1 DS235745 EEB16223.1 ADTU01024931 ADTU01024932 GL888120 EGI66989.1 ACPB03018955 ACPB03018956 CVRI01000063 CRL04336.1 KZ288349 PBC27405.1 KQ435916 KOX68775.1 KQ976719 KYM76749.1 KQ977329 KYN03554.1 KQ980159 KYN17432.1 KQ983089 KYQ47571.1 GL451091 EFN80055.1 GBYB01002243 JAG72010.1 GAMC01004382 JAC02174.1 GAMC01004381 JAC02175.1 GAKP01020538 JAC38414.1 GAKP01020536 JAC38416.1 AJVK01011653 GBXI01007005 JAD07287.1 LSYS01001520 OPJ88762.1 GDHF01020600 GDHF01008679 JAI31714.1 JAI43635.1 AKCR02000032 PKK25639.1 NEDP02004422 OWF45618.1 KL670502 KFW79471.1 LMAW01002702 KQK77914.1 KK510019 KFP62778.1 AGTO01011106 KK667148 KFQ12215.1 JRES01000438 KNC31167.1 MUZQ01000262 OWK53871.1 KA648760 AFP63389.1 KL409911 KFQ92699.1 KK943097 KFQ52251.1 ABQF01036981 ABQF01036982 ABQF01036983 ABQF01036984 ABQF01036985 ABQF01036986 ABQF01036987 KL215558 KFV65385.1 AKHW03001857 KYO40838.1 KK744609 KFP39779.1 KL522416 KFO90071.1 JXJN01021708 KL320193 KFP54724.1 KK373456 KFV44737.1 JW862077 AFO94594.1 AADN05000628 ADON01023591 ADON01023592 ADON01023593 ADON01023594 CCAG010010711 AGCU01056393 AGCU01056394 AGCU01056395 AGCU01056396 AGCU01056397 AGCU01056398 AGCU01056399 AGCU01056400 AGCU01056401 AGCU01056402 KK719561 KFO64327.1 CH902620 EDV31335.1 NNAY01000450 OXU28308.1 KK634753 KFV57741.1 JSUE03017698 JSUE03017699 JSUE03017700 JSUE03017701 CH940649 EDW63994.1 KK735702 KFR15429.1 LSMT01000064 PFX29522.1 CM001268 EHH25340.1 AABR07030942 AABR07030943 AQIB01145198 AQIB01145199 AQIB01145200 AQIB01145201 AQIB01145202 AQIB01145203 AQIB01145204 AQIB01145205 JH001222 EGW13206.1 AHAT01000593 AHAT01000594 AHAT01000595 JU336114 AFE79867.1 KL205697 KFV73089.1 JU470766 AFH27570.1
BABH01028025 BABH01028026 ODYU01010899 SOQ56293.1 NWSH01002295 PCG68635.1 KQ459606 KPI91343.1 AGBW02014611 OWR41346.1 KQ459896 KPJ19483.1 PYGN01000031 PSN56928.1 NEVH01000003 PNF44135.1 PNF44134.1 GEZM01031220 JAV84966.1 GEDC01002555 JAS34743.1 KQ971380 EEZ97256.1 GL438128 EFN69536.1 KB632350 ERL93229.1 KQ414642 KOC66771.1 KQ434869 KZC09295.1 GFTR01008328 JAW08098.1 KK107161 EZA56576.1 QOIP01000003 RLU25214.1 GBGD01000173 JAC88716.1 KQ981276 KYN43508.1 UFQT01000015 SSX17771.1 DS235745 EEB16223.1 ADTU01024931 ADTU01024932 GL888120 EGI66989.1 ACPB03018955 ACPB03018956 CVRI01000063 CRL04336.1 KZ288349 PBC27405.1 KQ435916 KOX68775.1 KQ976719 KYM76749.1 KQ977329 KYN03554.1 KQ980159 KYN17432.1 KQ983089 KYQ47571.1 GL451091 EFN80055.1 GBYB01002243 JAG72010.1 GAMC01004382 JAC02174.1 GAMC01004381 JAC02175.1 GAKP01020538 JAC38414.1 GAKP01020536 JAC38416.1 AJVK01011653 GBXI01007005 JAD07287.1 LSYS01001520 OPJ88762.1 GDHF01020600 GDHF01008679 JAI31714.1 JAI43635.1 AKCR02000032 PKK25639.1 NEDP02004422 OWF45618.1 KL670502 KFW79471.1 LMAW01002702 KQK77914.1 KK510019 KFP62778.1 AGTO01011106 KK667148 KFQ12215.1 JRES01000438 KNC31167.1 MUZQ01000262 OWK53871.1 KA648760 AFP63389.1 KL409911 KFQ92699.1 KK943097 KFQ52251.1 ABQF01036981 ABQF01036982 ABQF01036983 ABQF01036984 ABQF01036985 ABQF01036986 ABQF01036987 KL215558 KFV65385.1 AKHW03001857 KYO40838.1 KK744609 KFP39779.1 KL522416 KFO90071.1 JXJN01021708 KL320193 KFP54724.1 KK373456 KFV44737.1 JW862077 AFO94594.1 AADN05000628 ADON01023591 ADON01023592 ADON01023593 ADON01023594 CCAG010010711 AGCU01056393 AGCU01056394 AGCU01056395 AGCU01056396 AGCU01056397 AGCU01056398 AGCU01056399 AGCU01056400 AGCU01056401 AGCU01056402 KK719561 KFO64327.1 CH902620 EDV31335.1 NNAY01000450 OXU28308.1 KK634753 KFV57741.1 JSUE03017698 JSUE03017699 JSUE03017700 JSUE03017701 CH940649 EDW63994.1 KK735702 KFR15429.1 LSMT01000064 PFX29522.1 CM001268 EHH25340.1 AABR07030942 AABR07030943 AQIB01145198 AQIB01145199 AQIB01145200 AQIB01145201 AQIB01145202 AQIB01145203 AQIB01145204 AQIB01145205 JH001222 EGW13206.1 AHAT01000593 AHAT01000594 AHAT01000595 JU336114 AFE79867.1 KL205697 KFV73089.1 JU470766 AFH27570.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000245037
+ More
UP000235965 UP000192223 UP000007266 UP000000311 UP000030742 UP000053825 UP000076502 UP000005203 UP000053097 UP000279307 UP000078541 UP000009046 UP000005205 UP000007755 UP000015103 UP000183832 UP000242457 UP000053105 UP000078540 UP000078542 UP000078492 UP000075809 UP000008237 UP000095300 UP000092462 UP000190648 UP000053872 UP000242188 UP000053258 UP000051836 UP000016665 UP000037069 UP000197619 UP000095301 UP000053283 UP000007754 UP000092443 UP000085678 UP000053875 UP000091820 UP000050525 UP000189705 UP000078200 UP000092460 UP000092445 UP000000539 UP000016666 UP000001646 UP000092444 UP000007267 UP000002358 UP000052976 UP000007801 UP000215335 UP000006718 UP000001645 UP000008792 UP000053605 UP000189706 UP000225706 UP000079721 UP000002494 UP000029965 UP000001075 UP000018468 UP000053584
UP000235965 UP000192223 UP000007266 UP000000311 UP000030742 UP000053825 UP000076502 UP000005203 UP000053097 UP000279307 UP000078541 UP000009046 UP000005205 UP000007755 UP000015103 UP000183832 UP000242457 UP000053105 UP000078540 UP000078542 UP000078492 UP000075809 UP000008237 UP000095300 UP000092462 UP000190648 UP000053872 UP000242188 UP000053258 UP000051836 UP000016665 UP000037069 UP000197619 UP000095301 UP000053283 UP000007754 UP000092443 UP000085678 UP000053875 UP000091820 UP000050525 UP000189705 UP000078200 UP000092460 UP000092445 UP000000539 UP000016666 UP000001646 UP000092444 UP000007267 UP000002358 UP000052976 UP000007801 UP000215335 UP000006718 UP000001645 UP000008792 UP000053605 UP000189706 UP000225706 UP000079721 UP000002494 UP000029965 UP000001075 UP000018468 UP000053584
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
CDD
ProteinModelPortal
H9JSK3
A0A2H1WTB1
A0A2A4J981
A0A194PE89
A0A212EIN0
A0A194RNR9
+ More
A0A2P8ZKA7 A0A2J7RTG4 A0A2J7RTH2 A0A1Y1MM56 A0A1W4WQ67 A0A1B6E9Z8 D6X4J7 E2AAI9 U4UGW6 A0A0L7R7M2 A0A154PBN3 A0A088AJI0 A0A224XE77 A0A026WLQ2 A0A3L8DXL5 A0A069DY10 A0A195FST5 A0A336LLZ4 E0VS67 A0A158NSH8 F4WFL2 T1HW65 A0A1J1IW54 A0A2A3E6R3 A0A0M8ZQ24 A0A151HZF5 A0A195CTU5 A0A151J4C1 A0A151WI90 E2BW23 A0A0C9PME1 W8BZN8 W8CBY4 A0A034V7C1 A0A034V548 A0A1I8PLG6 A0A1B0D4Y0 A0A0A1X877 A0A1V4KWK5 A0A0K8VYF4 A0A2I0M7H9 A0A210QA62 A0A093PSQ2 A0A0Q3URE3 A0A091LTC1 U3JDB9 A0A091PX27 A0A0L0CG22 A0A218UJW0 A0A1I8M207 T1PKW7 A0A091UP09 A0A091S7W3 H0ZB90 A0A1A9X610 A0A1S3K523 A0A093G5C7 A0A1A9WDM9 A0A151NVM2 A0A1U7RZZ8 A0A1A9V6V7 A0A091KGS2 A0A091H2M9 A0A1B0BWC5 A0A091LEG0 A0A093EU46 V9K9Q8 A0A1B0A5Y4 A0A1D5NTW7 E1BU18 U3ITR5 G1KU24 A0A1B0FQX1 K7GD88 K7IW10 A0A091F635 B3MJC5 A0A232FCX9 A0A093FTT0 F7GUM4 G1MX06 B4LTU3 A0A091WIR9 A0A3Q0CJ53 A0A2B4SLY4 A0A1S3WCX2 G7NHJ1 F1M1D5 A0A0D9S509 G3I449 W5N9F6 H9FZ08 A0A093J268 H9YV54
A0A2P8ZKA7 A0A2J7RTG4 A0A2J7RTH2 A0A1Y1MM56 A0A1W4WQ67 A0A1B6E9Z8 D6X4J7 E2AAI9 U4UGW6 A0A0L7R7M2 A0A154PBN3 A0A088AJI0 A0A224XE77 A0A026WLQ2 A0A3L8DXL5 A0A069DY10 A0A195FST5 A0A336LLZ4 E0VS67 A0A158NSH8 F4WFL2 T1HW65 A0A1J1IW54 A0A2A3E6R3 A0A0M8ZQ24 A0A151HZF5 A0A195CTU5 A0A151J4C1 A0A151WI90 E2BW23 A0A0C9PME1 W8BZN8 W8CBY4 A0A034V7C1 A0A034V548 A0A1I8PLG6 A0A1B0D4Y0 A0A0A1X877 A0A1V4KWK5 A0A0K8VYF4 A0A2I0M7H9 A0A210QA62 A0A093PSQ2 A0A0Q3URE3 A0A091LTC1 U3JDB9 A0A091PX27 A0A0L0CG22 A0A218UJW0 A0A1I8M207 T1PKW7 A0A091UP09 A0A091S7W3 H0ZB90 A0A1A9X610 A0A1S3K523 A0A093G5C7 A0A1A9WDM9 A0A151NVM2 A0A1U7RZZ8 A0A1A9V6V7 A0A091KGS2 A0A091H2M9 A0A1B0BWC5 A0A091LEG0 A0A093EU46 V9K9Q8 A0A1B0A5Y4 A0A1D5NTW7 E1BU18 U3ITR5 G1KU24 A0A1B0FQX1 K7GD88 K7IW10 A0A091F635 B3MJC5 A0A232FCX9 A0A093FTT0 F7GUM4 G1MX06 B4LTU3 A0A091WIR9 A0A3Q0CJ53 A0A2B4SLY4 A0A1S3WCX2 G7NHJ1 F1M1D5 A0A0D9S509 G3I449 W5N9F6 H9FZ08 A0A093J268 H9YV54
Ontologies
GO
GO:0007021
GO:0007023
GO:0005096
GO:0048487
GO:0016021
GO:0006457
GO:0070830
GO:0005923
GO:0000226
GO:0005912
GO:0034333
GO:0016328
GO:0000278
GO:0005813
GO:0048667
GO:0031115
GO:0006352
GO:0005634
GO:1902850
GO:0045196
GO:0007409
GO:0048813
GO:0010842
GO:0007420
GO:0005488
GO:0007186
GO:0016020
GO:0004888
GO:0007166
GO:0016616
GO:0046168
GO:0016491
GO:0003824
PANTHER
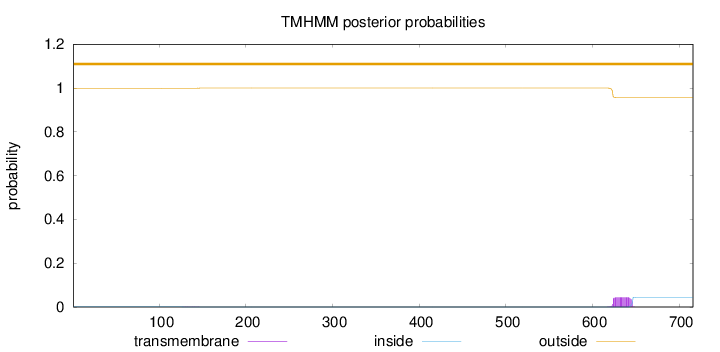
Topology
Length:
716
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00602
Exp number, first 60 AAs:
0.00092
Total prob of N-in:
0.00079
outside
1 - 716
Population Genetic Test Statistics
Pi
263.230475
Theta
177.749148
Tajima's D
1.423708
CLR
0
CSRT
0.767511624418779
Interpretation
Uncertain
